Nasopharyngeal microbiota in hospitalized children with Bordetella pertussis and Rhinovirus infection
- PMID: 34819600
- PMCID: PMC8613181
- DOI: 10.1038/s41598-021-02322-y
Nasopharyngeal microbiota in hospitalized children with Bordetella pertussis and Rhinovirus infection
Abstract
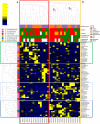
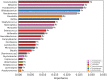
Despite great advances in describing Bordetella pertussis infection, the role of the host microbiota in pertussis pathogenesis remains unexplored. Indeed, the microbiota plays important role in defending against bacterial and viral respiratory infections. We investigated the nasopharyngeal microbiota in infants infected by B. pertussis (Bp), Rhinovirus (Rv) and simultaneously by both infectious agents (Bp + Rv). We demonstrated a specific nasopharyngeal microbiome profiles for Bp group, compared to Rv and Bp + Rv groups, and a reduction of microbial richness during coinfection compared to the single infections. The comparison amongst the three groups showed the increase of Alcaligenaceae and Achromobacter in Bp and Moraxellaceae and Moraxella in Rv group. Furthermore, correlation analysis between patients' features and nasopharyngeal microbiota profile highlighted a link between delivery and feeding modality, antibiotic administration and B. pertussis infection. A model classification demonstrated a microbiota fingerprinting specific of Bp and Rv infections. In conclusion, external factors since the first moments of life contribute to the alteration of nasopharyngeal microbiota, indeed increasing the susceptibility of the host to the pathogens' infections. When the infection is triggered, the presence of infectious agents modifies the microbiota favoring the overgrowth of commensal bacteria that turn in pathobionts, hence contributing to the disease severity.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Iorio, A. et al. Cross-correlation of virome–bacteriome–host–metabolome to study respiratory health. Trends Microbiol. S0966842X21001220 (2021) 10.1016/j.tim.2021.04.011. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

