The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer
- PMID: 34819663
- PMCID: PMC8687755
- DOI: 10.1038/s41586-021-04135-5
The CLIP1-LTK fusion is an oncogenic driver in non-small-cell lung cancer
Abstract
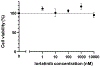
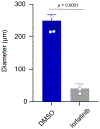
Lung cancer is one of the most aggressive tumour types. Targeted therapies stratified by oncogenic drivers have substantially improved therapeutic outcomes in patients with non-small-cell lung cancer (NSCLC)1. However, such oncogenic drivers are not found in 25-40% of cases of lung adenocarcinoma, the most common histological subtype of NSCLC2. Here we identify a novel fusion transcript of CLIP1 and LTK using whole-transcriptome sequencing in a multi-institutional genome screening platform (LC-SCRUM-Asia, UMIN000036871). The CLIP1-LTK fusion was present in 0.4% of NSCLCs and was mutually exclusive with other known oncogenic drivers. We show that kinase activity of the CLIP1-LTK fusion protein is constitutively activated and has transformation potential. Treatment of Ba/F3 cells expressing CLIP1-LTK with lorlatinib, an ALK inhibitor, inhibited CLIP1-LTK kinase activity, suppressed proliferation and induced apoptosis. One patient with NSCLC harbouring the CLIP1-LTK fusion showed a good clinical response to lorlatinib treatment. To our knowledge, this is the first description of LTK alterations with oncogenic activity in cancers. These results identify the CLIP1-LTK fusion as a target in NSCLC that could be treated with lorlatinib.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures





Comment in
-
LTK fusions: A new target emerges in non-small cell lung cancer.Cancer Cell. 2022 Jan 10;40(1):23-25. doi: 10.1016/j.ccell.2021.12.012. Cancer Cell. 2022. PMID: 35016028 Free PMC article.
-
The CLIP1-LTK fusion: a new oncogenic driver in non-small-cell lung cancer?Future Oncol. 2023 Feb;19(4):273-275. doi: 10.2217/fon-2022-1232. Epub 2023 Mar 21. Future Oncol. 2023. PMID: 36942735 No abstract available.
References
-
- Fernandez-Cuesta L et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discov 4, 415–422, doi: 10.1158/2159-8290.CD-13-0633 (2014). - DOI - PubMed
-
- National Comprehensive Cancer Network. Non-Small Cell Lung Cancer (Version 4.2021), <https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf> (2021).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

