Genome-Wide Analysis of CCT Transcript Factors to Identify Genes Contributing to Photoperiodic Flowering in Oryza rufipogon
- PMID: 34819938
- PMCID: PMC8606741
- DOI: 10.3389/fpls.2021.736419
Genome-Wide Analysis of CCT Transcript Factors to Identify Genes Contributing to Photoperiodic Flowering in Oryza rufipogon
Abstract
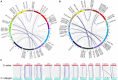
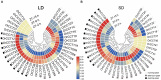
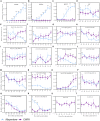
Photoperiod sensitivity is a dominant determinant for the phase transition in cereal crops. CCT (CONSTANS, CO-like, and TOC1) transcription factors (TFs) are involved in many physiological functions including the regulation of the photoperiodic flowering. However, the functional roles of CCT TFs have not been elucidated in the wild progenitors of crops. In this study, we identified 41 CCT TFs, including 19 CMF, 17 COL, and five PRR TFs in Oryza rufipogon, the presumed wild ancestor of Asian cultivated rice. There are thirty-eight orthologous CCT genes in Oryza sativa, of which ten pairs of duplicated CCT TFs are shared with O. rufipogon. We investigated daily expression patterns, showing that 36 OrCCT genes exhibited circadian rhythmic expression. A total of thirteen OrCCT genes were identified as putative flowering suppressors in O. rufipogon based on rhythmic and developmental expression patterns and transgenic phenotypes. We propose that OrCCT08, OrCCT24, and OrCCT26 are the strong functional alleles of rice DTH2, Ghd7, and OsPRR37, respectively. The SD treatment at 80 DAG stimulated flowering of the LD-grown O. rufipogon plants. Our results further showed that the nine OrCCT genes were significantly downregulated under the treatment. Our findings would provide valuable information for the construction of photoperiodic flowering regulatory network and functional characterization of the CCT TFs in both O. rufipogon and O. sativa.
Keywords: CCT genes; Oryza rufipogon; expression profiles; genomic synteny; photoperiodic flowering regulation; rice.
Copyright © 2021 Peng, Tun, Dai, Li, Zhang, Yin, Yoon, Cho, An and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- An G., Ebert P. R., Mitra A., Ha S. B. (1989). “Binary vectors,” in Plant Molecular Biology Manual, ed. Kluwer Academic Publisher (Dordrecht: Springer; ), 29–47.
LinkOut - more resources
Full Text Sources
Research Materials

