Direct detection of intact Klebsiella pneumoniae carbapenemase variants from cell lysates: Identification, characterization and clinical implications
- PMID: 34820520
- PMCID: PMC8600995
- DOI: 10.1016/j.clinms.2020.07.001
Direct detection of intact Klebsiella pneumoniae carbapenemase variants from cell lysates: Identification, characterization and clinical implications
Abstract
Introduction: Carbapenemase-producing organisms (CPOs) are a growing threat to human health. Among the enzymes conferring antibiotic resistance produced by these organisms, Klebsiella pneumoniae carbapenemase (KPC) is considered to be a growing global health threat. Reliable and specific detection of this antibiotic resistance-causing enzyme is critical both for effective therapy and to mitigate further spread.
Objectives: The objective of this study is to develop an intact protein mass spectrometry-based method for detection and differentiation of clinically-relevant KPC variants directly from bacterial cell lysates. The method should be specific for any variant expressed in multiple bacterial species, limit false positive results and be rapid in nature to directly influence clinical outcomes.
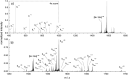
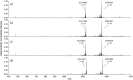
Methods: Lysates obtained directly from bacterial colonies were used for intact protein detection using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Bottom-up and top-down proteomic methods were used to characterize the KPC protein targets of interest. Comparisons between KPC-producing and KPC-non-producing isolates from a wide variety of species were also performed.
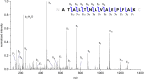
Results: Characterization of the mature KPC protein revealed an unexpected signal peptide cleavage site preceding an AXA signal peptide motif, modifying the molecular weight (MW) of the mature protein. Taking the additional AXA residues into account allowed for direct detection of the intact protein using top-down proteomic methods. Further validation was performed by transforming a KPC-harboring plasmid into a negative control strain, followed by MS detection of the KPC variant from the transformed cell line. Application of this approach to clearly identify clinically-relevant variants among several species is presented for KPC-2, KPC-3, KPC-4 and KPC-5.
Conclusion: Direct detection of these enzymes contributes to the understanding of occurrence and spread of these antibiotic-resistant organisms. The ability to detect intact KPC variants via a simple LC-MS/MS approach could have a direct and positive impact on clinical therapy, by providing both direction for epidemiological tracking and appropriate therapy.
Keywords: ATCC, American type culture collection; BLAST, basic local alignment search tool; CDC, Centers for Disease Control and Prevention; CPO, carbapenemase-producing organisms; CSD, charge state distribution; Carbapenem-resistant Enterobacteriaceae; Carbapenemase-producing organisms; ESI, electrospray ionization; KPC, Klebsiella pneumoniae carbapenemase; Klebsiella pneumoniae carbapenemase; LC, liquid chromatography; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; MS/MS, tandem mass spectrometry; MW, molecular weight; Mass Spectrometry; PCR, polymerase chain reaction; TOF, time-of-flight; Tandem mass spectrometry; m/z, mass-to-charge ratio.
© 2020 The Association for Mass Spectrometry: Applications to the Clinical Lab (MSACL). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
W. M. McGee, J. R. Neil, S. R. Kronewitter, and J. L. Stephenson, Jr. are employees of Thermo Fisher Scientific. Also, W.M. McGee, J.R. Neil, and J.L. Stephenson Jr. own stock in Thermo Fisher Scientific. There are two patents that have been filed on resistance marker identification by the authors from Thermo Fisher Scientific. M. L. Faron, B. W. Buchan and N. A. Ledeboer have received research funding from Bruker Daltonics and bioMerieux, and N. A. Ledeboer is a consultant for Thermo Fisher Scientific.
Figures


References
-
- Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heuer O.E., Kahlmeter G., Kruse H., Laxminarayan R., Liebana E., Lopez-Cerero L., MacGowan A., Martins M., Rodriguez-Bano J., Rolain J.M., Segovia C., Sigauque B., Tacconelli E., Wellington E., Vila J. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infections. 2015;6:22–29. - PMC - PubMed
-
- Antibiotic C.D.C. Services, CDC; 2013. Resistance Threats in the United States. Atlanta, GA: U.S. Department of Health and Human.
-
- CDC. Antibiotic Resistance threats in the United States. Atlanta, GA: U. S. Department of Health and Human Services, CDC, 2019.
LinkOut - more resources
Full Text Sources
Medical
Research Materials
