A cryptic variation in a member of the Ovate Family Proteins is underlying the melon fruit shape QTL fsqs8.1
- PMID: 34821982
- PMCID: PMC8942903
- DOI: 10.1007/s00122-021-03998-6
A cryptic variation in a member of the Ovate Family Proteins is underlying the melon fruit shape QTL fsqs8.1
Abstract
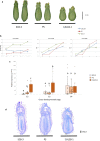
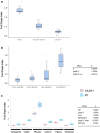
The gene underlying the melon fruit shape QTL fsqs8.1 is a member of the Ovate Family Proteins. Variation in fruit morphology is caused by changes in gene expression likely due to a cryptic structural variation in this locus. Melon cultivars have a wide range of fruit morphologies. Quantitative trait loci (QTL) have been identified underlying such diversity. This research focuses on the fruit shape QTL fsqs8.1, previously detected in a cross between the accession PI 124112 (CALC, producing elongated fruit) and the cultivar 'Piel de Sapo' (PS, producing oval fruit). The CALC fsqs8.1 allele induced round fruit shape, being responsible for the transgressive segregation for this trait observed in that population. In fact, the introgression line CALC8-1, carrying the fsqs8.1 locus from CALC into the PS genetic background, produced perfect round fruit. Following a map-based cloning approach, we found that the gene underlying fsqs8.1 is a member of the Ovate Family Proteins (OFP), CmOFP13, likely a homologue of AtOFP1 and SlOFP20 from Arabidopsis thaliana and tomato, respectively. The induction of the round shape was due to the higher expression of the CALC allele at the early ovary development stage. The fsqs8.1 locus showed an important structural variation, being CmOFP13 surrounded by two deletions in the CALC genome. The deletions are present at very low frequency in melon germplasm. Deletions and single nucleotide polymorphisms in the fsqs8.1 locus could not be not associated with variation in fruit shape among different melon accessions, what indicates that other genetic factors should be involved to induce the CALC fsqs8.1 allele effects. Therefore, fsqs8.1 is an example of a cryptic variation that alters gene expression, likely due to structural variation, resulting in phenotypic changes in melon fruit morphology.
© 2021. The Author(s).
Conflict of interest statement
Authors declare no conflict of interest.
Figures




Similar articles
-
Mapping and introgression of QTL involved in fruit shape transgressive segregation into ‘piel de sapo’ melon (cucumis melo l.) [corrected].PLoS One. 2014 Aug 15;9(8):e104188. doi: 10.1371/journal.pone.0104188. eCollection 2014. PLoS One. 2014. PMID: 25126852 Free PMC article.
-
CmFSI8/CmOFP13 encoding an OVATE family protein controls fruit shape in melon.J Exp Bot. 2022 Mar 2;73(5):1370-1384. doi: 10.1093/jxb/erab510. J Exp Bot. 2022. PMID: 34849737
-
The genetic basis of fruit morphology in horticultural crops: lessons from tomato and melon.J Exp Bot. 2014 Aug;65(16):4625-37. doi: 10.1093/jxb/eru017. Epub 2014 Feb 11. J Exp Bot. 2014. PMID: 24520021 Review.
-
A Novel Introgression Line Library Derived from a Wild Melon Gives Insights into the Genetics of Melon Domestication, Uncovering New Genetic Variability Useful for Breeding.Int J Mol Sci. 2023 Jun 14;24(12):10099. doi: 10.3390/ijms241210099. Int J Mol Sci. 2023. PMID: 37373247 Free PMC article.
-
Genetic architecture of fruit size and shape variation in cucurbits: a comparative perspective.Theor Appl Genet. 2020 Jan;133(1):1-21. doi: 10.1007/s00122-019-03481-3. Epub 2019 Nov 25. Theor Appl Genet. 2020. PMID: 31768603 Review.
Cited by
-
Fruit Morphology and Ripening-Related QTLs in a Newly Developed Introgression Line Collection of the Elite Varieties 'Védrantais' and 'Piel de Sapo'.Plants (Basel). 2022 Nov 15;11(22):3120. doi: 10.3390/plants11223120. Plants (Basel). 2022. PMID: 36432848 Free PMC article.
-
Pan-genome and multi-parental framework for high-resolution trait dissection in melon (Cucumis melo).Plant J. 2022 Dec;112(6):1525-1542. doi: 10.1111/tpj.16021. Epub 2022 Nov 23. Plant J. 2022. PMID: 36353749 Free PMC article.
-
A Rare Frameshift Mutation of in CmACS7 Alters Ethylene Biosynthesis and Determines Fruit Morphology in Melon (Cucumis melo L.).Plants (Basel). 2025 Jul 8;14(14):2087. doi: 10.3390/plants14142087. Plants (Basel). 2025. PMID: 40733324 Free PMC article.
-
Genomic analysis of fruit size and shape traits in apple: unveiling candidate genes through GWAS analysis.Hortic Res. 2023 Dec 19;11(2):uhad270. doi: 10.1093/hr/uhad270. eCollection 2024 Feb. Hortic Res. 2023. PMID: 38419968 Free PMC article.
-
Genome-wide analysis of OFP gene family in pepper (Capsicum annuum L.).Front Genet. 2022 Sep 30;13:941954. doi: 10.3389/fgene.2022.941954. eCollection 2022. Front Genet. 2022. PMID: 36246640 Free PMC article.
References
-
- Abdelmohsin ME, Pitrat M (2008) Pleiotropic effect of sex expression on fruit shape in melon. In: Pitrat M (ed) 9th EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, INRA, Avignon, France, pp 551–555
-
- Alonge M, Wang XG, Benoit M, Soyk S, Pereira L, Zhang L, Suresh H, Ramakrishnan S, Maumus F, Ciren D, Levy Y, Harel TH, Shalev-Schlosser G, Amsellem Z, Razifard H, Caicedo AL, Tieman DM, Klee H, Kirsche M, Aganezov S, Ranallo-Benavidez TR, Lemmon ZH, Kim J, Robitaille G, Kramer M, Goodwin S, McCombie WR, Hutton S, Van Eck J, Gillis J, Eshed Y, Sedlazeck FJ, van der Knaap E, Schatz MC, Lippman ZB. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182:145–161. - PMC - PubMed
-
- Bai SL , Peng YB, Cui JX, Gu HT, Xu LY, Li YQ, Xu ZH, Bai SN (2004). Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.) Planta 220: 230–240 - PubMed
-
- Boualem A, Fergany M, Fernandez R, Troadec C, Martin A, Morin H, Sari MA, Collin F, Flowers JM, Pitrat M, Purugganan MD, Dogimont C, Bendahmane A. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science. 2008;321:836–838. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

