Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories
- PMID: 34822296
- PMCID: PMC8896201
- DOI: 10.1126/science.abh2444
Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories
Abstract
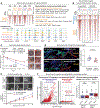
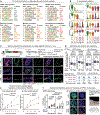
Immune and tissue stem cells retain an epigenetic memory of inflammation that intensifies sensitivity to future encounters. We investigated whether and to what consequence stem cells possess and accumulate memories of diverse experiences. Monitoring a choreographed response to wounds, we found that as hair follicle stem cells leave their niche, migrate to repair damaged epidermis, and take up long-term foreign residence there, they accumulate long-lasting epigenetic memories of each experience, culminating in post-repair epigenetic adaptations that sustain the epidermal transcriptional program and surface barrier. Each memory is distinct, separable, and has its own physiological impact, collectively endowing these stem cells with heightened regenerative ability to heal wounds and broadening their tissue-regenerating tasks relative to their naïve counterparts.
Conflict of interest statement
Figures



Comment in
-
Stem cells remember insults.Science. 2021 Nov 26;374(6571):1052-1053. doi: 10.1126/science.abm6806. Epub 2021 Nov 25. Science. 2021. PMID: 34822297
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

