Inhibition of USP14 influences alphaherpesvirus proliferation by degrading viral VP16 protein via ER stress-triggered selective autophagy
- PMID: 34822318
- PMCID: PMC9450976
- DOI: 10.1080/15548627.2021.2002101
Inhibition of USP14 influences alphaherpesvirus proliferation by degrading viral VP16 protein via ER stress-triggered selective autophagy
Abstract
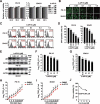
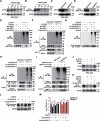
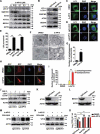
Alphaherpesvirus infection results in severe health consequences in a wide range of hosts. USPs are the largest subfamily of deubiquitinating enzymes that play critical roles in immunity and other cellular functions. To investigate the role of USPs in alphaherpesvirus replication, we assessed 13 USP inhibitors for PRV replication. Our data showed that all the tested compounds inhibited PRV replication, with the USP14 inhibitor b-AP15 exhibiting the most dramatic effect. Ablation of USP14 also influenced PRV replication, whereas replenishment of USP14 in USP14 null cells restored viral replication. Although inhibition of USP14 induced the K63-linked ubiquitination of PRV VP16 protein, its degradation was not dependent on the proteasome. USP14 directly bound to ubiquitin chains on VP16 through its UBL domain during the early stage of viral infection. Moreover, USP14 inactivation stimulated EIF2AK3/PERK- and ERN1/IRE1-mediated signaling pathways, which were responsible for VP16 degradation through SQSTM1/p62-mediated selective macroautophagy/autophagy. Ectopic expression of non-ubiquitinated VP16 fully rescued PRV replication. Challenge of mice with b-AP15 activated ER stress and autophagy and inhibited PRV infection in vivo. Our results suggested that USP14 was a potential therapeutic target to treat alphaherpesvirus-induced infectious diseases.Abbreviations ATF4: activating transcription factor 4; ATF6: activating transcription factor 6; ATG5: autophagy related 5; ATG12: autophagy related 12; CCK-8: cell counting kit-8; Co-IP: co-immunoprecipitation; CRISPR: clustered regulatory interspaced short palindromic repeat; Cas9: CRISPR associated system 9; DDIT3/CHOP: DNA-damage inducible transcript 3; DNAJB9/ERdj4: DnaJ heat shock protein family (Hsp40) member B9; DUBs: deubiquitinases; EIF2A/eIF2α: eukaryotic translation initiation factor 2A; EIF2AK3/PERK: eukaryotic translation initiation factor 2 alpha kinase 3; EP0: ubiquitin E3 ligase ICP0; ER: endoplasmic reticulum; ERN1/IRE1: endoplasmic reticulum (ER) to nucleus signaling 1; FOXO1: forkhead box O1; FRET: Förster resonance energy transfer; HSPA5/BiP: heat shock protein 5; HSV: herpes simplex virus; IE180: transcriptional regulator ICP4; MAP1LC3/LC3: microtube-associated protein 1 light chain 3; MOI: multiplicity of infection; MTOR: mechanistic target of rapamycin kinase; PPP1R15A/GADD34: protein phosphatase 1, regulatory subunit 15A; PRV: pseudorabies virus; PRV gB: PRV glycoprotein B; PRV gE: PRV glycoprotein E; qRT-PCR: quantitative real-time polymerase chain reaction; sgRNA: single guide RNA; siRNA: small interfering RNA; SQSTM1/p62: sequestosome 1; TCID50: tissue culture infective dose; UB: ubiquitin; UBA: ubiquitin-associated domain; UBL: ubiquitin-like domain; UL9: DNA replication origin-binding helicase; UPR: unfolded protein response; USPs: ubiquitin-specific proteases; VHS: virion host shutoff; VP16: viral protein 16; XBP1: X-box binding protein 1; XBP1s: small XBP1; XBP1(t): XBP1-total.
Keywords: Alphaherpesvirus; ER stress; PRV VP16; USP14; selective autophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
-
- Bharucha T, Houlihan CF, Breuer J, et al. Herpesvirus infections of the central nervous system. Semin Neurol. 2019;39(3):369–382. - PubMed
-
- Koshizuka T, Suzutani T.. [Anti alpha-herpesvirus drugs]. Nihon Rinsho. 2012;70:558–563. - PubMed
-
- Muller T, Hahn EC, Tottewitz F, et al. Pseudorabies virus in wild swine: a global perspective. Arch Virol. 2011;156(10):1691–1705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
