Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2
- PMID: 34822776
- PMCID: PMC8590493
- DOI: 10.1016/j.chom.2021.11.005
Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2
Abstract
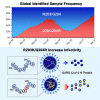
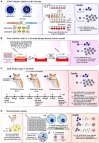
Previous work found that the co-occurring mutations R203K/G204R on the SARS-CoV-2 nucleocapsid (N) protein are increasing in frequency among emerging variants of concern or interest. Through a combination of in silico analyses, this study demonstrates that R203K/G204R are adaptive, while large-scale phylogenetic analyses indicate that R203K/G204R associate with the emergence of the high-transmissibility SARS-CoV-2 lineage B.1.1.7. Competition experiments suggest that the 203K/204R variants possess a replication advantage over the preceding R203/G204 variants, possibly related to ribonucleocapsid (RNP) assembly. Moreover, the 203K/204R virus shows increased infectivity in human lung cells and hamsters. Accordingly, we observe a positive association between increased COVID-19 severity and sample frequency of 203K/204R. Our work suggests that the 203K/204R mutations contribute to the increased transmission and virulence of select SARS-CoV-2 variants. In addition to mutations in the spike protein, mutations in the nucleocapsid protein are important for viral spreading during the pandemic.
Keywords: COVID-19; R203K/G204R; SARS-CoV-2; fatality; fitness; infectivity; mutation; nucleocapsid; virulence.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

