Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation
- PMID: 34823256
- PMCID: PMC8828475
- DOI: 10.1038/s41586-021-04266-9
Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation
Abstract
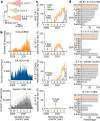
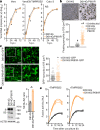
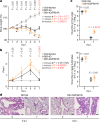
During the current coronavirus disease 2019 (COVID-19) pandemic, a variety of mutations have accumulated in the viral genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and, at the time of writing, four variants of concern are considered to be potentially hazardous to human society1. The recently emerged B.1.617.2/Delta variant of concern is closely associated with the COVID-19 surge that occurred in India in the spring of 2021 (ref. 2). However, the virological properties of B.1.617.2/Delta remain unclear. Here we show that the B.1.617.2/Delta variant is highly fusogenic and notably more pathogenic than prototypic SARS-CoV-2 in infected hamsters. The P681R mutation in the spike protein, which is highly conserved in this lineage, facilitates cleavage of the spike protein and enhances viral fusogenicity. Moreover, we demonstrate that the P681R-bearing virus exhibits higher pathogenicity compared with its parental virus. Our data suggest that the P681R mutation is a hallmark of the virological phenotype of the B.1.617.2/Delta variant and is associated with enhanced pathogenicity.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Coronavirus Disease 2019 (WHO, 2021); https://www.who.int/emergencies/diseases/novel-coronavirus-2019
-
- Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. - PubMed
-
- Zhou B, et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

