Comparative and phylogenetic analyses of eleven complete chloroplast genomes of Dipterocarpoideae
- PMID: 34823565
- PMCID: PMC8620154
- DOI: 10.1186/s13020-021-00538-8
Comparative and phylogenetic analyses of eleven complete chloroplast genomes of Dipterocarpoideae
Abstract
Background: In South-east Asia, Dipterocarpoideae is predominant in most mature forest communities, comprising around 20% of all trees. As large quantity and high quality wood are produced in many species, Dipterocarpoideae plants are the most important and valuable source in the timber market. The d-borneol is one of the essential oil components from Dipterocarpoideae (for example, Dryobalanops aromatica or Dipterocarpus turbinatus) and it is also an important traditional Chinese medicine (TCM) formulation known as "Bingpian" in Chinese, with antibacterial, analgesic and anti-inflammatory effects and can enhance anticancer efficiency.
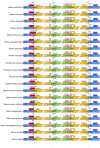
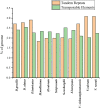
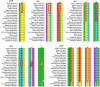
Methods: In this study, we analyzed 20 chloroplast (cp) genomes characteristics of Dipterocarpoideae, including eleven newly reported genomes and nine cp genomes previously published elsewhere, then we explored the chloroplast genomic features, inverted repeats contraction and expansion, codon usage, amino acid frequency, the repeat sequences and selective pressure analyses. At last, we constructed phylogenetic relationships of Dipterocarpoideae and found the potential barcoding loci.
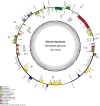
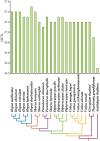
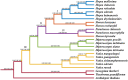
Results: The cp genome of this subfamily has a typical quadripartite structure and maintains a high degree of consistency among species. There were slightly more tandem repeats in cp genomes of Dipterocarpus and Vatica, and the psbH gene was subjected to positive selection in the common ancestor of all the 20 species of Dipterocarpoideae compared with three outgroups. Phylogenetic tree showed that genus Shorea was not a monophyletic group, some Shorea species and genus Parashorea are placed in one clade. In addition, the rpoC2 gene can be used as a potential marker to achieve accurate and rapid species identification in subfamily Dipterocarpoideae.
Conclusions: Dipterocarpoideae had similar cp genomic features and psbM, rbcL, psbH may function in the growth of Dipterocarpoideae. Phylogenetic analysis suggested new taxon treatment is needed for this subfamily indentification. In addition, rpoC2 is potential to be a barcoding gene to TCM distinguish.
Keywords: Chloroplast genomes; Comparative genomics; DNA barcoding; Dipterocarpoideae; Phylogenetics; Selected selection.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Brearley FQ, Banin LF, Saner P. The ecology of the Asian dipterocarps. Plant Ecol Divers. 2017;9(5-6):429–436.
-
- Ashton PS. Dipterocarpaceae. vol. 9; 1982.
-
- Dwiyanti FG, Kamiya K, Harada K. Phylogeographic structure of the commercially important tropical tree species, Dryobalanops aromatica gaertn. f. (Dipterocarpaceae) revealed by microsatellite markers. Reinwardtia. 2014;14(1):43–51.
-
- Slik JWF, Poulsen AD, Ashton PS, Cannon CH, Eichhorn KAO, Kartawinata K, Lanniari I, Nagamasu H, Nakagawa M, Van Nieuwstadt MGL, et al. A floristic analysis of the lowland dipterocarp forests of Borneo. J Biogeogr. 2003;30(10):1517–1531.
-
- Appanah S, Turnbull JM. A review of dipterocarps: Taxonomy, ecology, and silviculture. 1998.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

