NogoA-expressing astrocytes limit peripheral macrophage infiltration after ischemic brain injury in primates
- PMID: 34824275
- PMCID: PMC8617297
- DOI: 10.1038/s41467-021-27245-0
NogoA-expressing astrocytes limit peripheral macrophage infiltration after ischemic brain injury in primates
Abstract
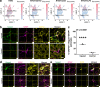
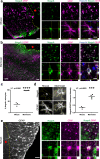
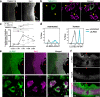
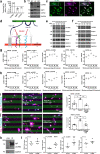
Astrocytes play critical roles after brain injury, but their precise function is poorly defined. Utilizing single-nuclei transcriptomics to characterize astrocytes after ischemic stroke in the visual cortex of the marmoset monkey, we observed nearly complete segregation between stroke and control astrocyte clusters. Screening for the top 30 differentially expressed genes that might limit stroke recovery, we discovered that a majority of astrocytes expressed RTN4A/ NogoA, a neurite-outgrowth inhibitory protein previously only associated with oligodendrocytes. NogoA upregulation on reactive astrocytes post-stroke was significant in both the marmoset and human brain, whereas only a marginal change was observed in mice. We determined that NogoA mediated an anti-inflammatory response which likely contributes to limiting the infiltration of peripheral macrophages into the surviving parenchyma.
© 2021. The Author(s).
Conflict of interest statement
S.M.S. is a founder and equity holder in ReNetX Bio, Inc., which seeks clinical development of NgR1-FC (AXER-204) for spinal cord injury. The remaining authors declare no competing interests.
Figures

References
-
- O’collins VE, et al. 1,026 experimental treatments in acute stroke. Ann. Neurol. 2006;59:467–477. - PubMed
-
- Liddelow SA, Sofroniew MV. Astrocytes usurp neurons as a disease focus. Nat. Neurosci. 2019;22:512–513. - PubMed
-
- Liddelow SA, Barres BA. Reactive astrocytes: production, function, and therapeutic potential. Immunity. 2017;46:957–967. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

