Rank-invariant estimation of inbreeding coefficients
- PMID: 34824382
- PMCID: PMC8733021
- DOI: 10.1038/s41437-021-00471-4
Rank-invariant estimation of inbreeding coefficients
Abstract
The two alleles an individual carries at a locus are identical by descent (ibd) if they have descended from a single ancestral allele in a reference population, and the probability of such identity is the inbreeding coefficient of the individual. Inbreeding coefficients can be predicted from pedigrees with founders constituting the reference population, but estimation from genetic data is not possible without data from the reference population. Most inbreeding estimators that make explicit use of sample allele frequencies as estimates of allele probabilities in the reference population are confounded by average kinships with other individuals. This means that the ranking of those estimates depends on the scope of the study sample and we show the variation in rankings for common estimators applied to different subdivisions of 1000 Genomes data. Allele-sharing estimators of within-population inbreeding relative to average kinship in a study sample, however, do have invariant rankings across all studies including those individuals. They are unbiased with a large number of SNPs. We discuss how allele sharing estimates are the relevant quantities for a range of empirical applications.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



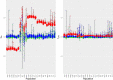

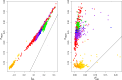
References
-
- Allen N, et al. UK Biobank: current status and what it means for epidemiology. Health Policy Technol. 2012;1:123–126.
-
- Astle W, Balding DJ. Population structure and cryptic relatedness in genetic association studies. Stat Sci. 2009;24:451–471.
-
- Ayres KL, Balding DJ. Measuring departures from Hardy-Weinberg: a Markov chain Monte Carlo method for estimating the inbreeding coefficient. Heredity. 1998;80:769–777. - PubMed
-
- Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. 2018;19:220–234. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

