Keratoconus Severity Classification Using Features Selection and Machine Learning Algorithms
- PMID: 34824602
- PMCID: PMC8610665
- DOI: 10.1155/2021/9979560
Keratoconus Severity Classification Using Features Selection and Machine Learning Algorithms
Abstract
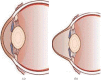
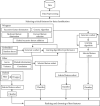
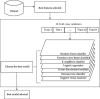
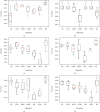
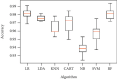
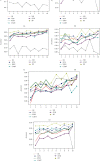
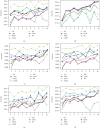
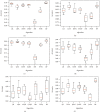
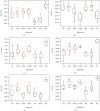
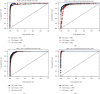
Keratoconus is a noninflammatory disease characterized by thinning and bulging of the cornea, generally appearing during adolescence and slowly progressing, causing vision impairment. However, the detection of keratoconus remains difficult in the early stages of the disease because the patient does not feel any pain. Therefore, the development of a method for detecting this disease based on machine and deep learning methods is necessary for early detection in order to provide the appropriate treatment as early as possible to patients. Thus, the objective of this work is to determine the most relevant parameters with respect to the different classifiers used for keratoconus classification based on the keratoconus dataset of Harvard Dataverse. A total of 446 parameters are analyzed out of 3162 observations by 11 different feature selection algorithms. Obtained results showed that sequential forward selection (SFS) method provided a subset of 10 most relevant variables, thus, generating the highest classification performance by the application of random forest (RF) classifier, with an accuracy of 98% and 95% considering 2 and 4 keratoconus classes, respectively. Found classification accuracy applying RF classifier on the selected variables using SFS method achieves the accuracy obtained using all features of the original dataset.
Copyright © 2021 Mustapha Aatila et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Figures








References
-
- Adnan K., Akbar R. An analytical study of information extraction from unstructured and multidimensional big data. Journal of Big Data . 2019;6(1) doi: 10.1186/s40537-019-0254-8. - DOI
-
- Reddy G. T., Reddy M. P. K., Lakshmanna K., et al. Analysis of dimensionality reduction techniques on big data. IEEE Access . 2020;8:54776–54788. doi: 10.1109/ACCESS.2020.2980942. - DOI
-
- Obaid H. S., Dheyab S. A., Sabry S. S. The impact of data pre-processing techniques and dimensionality reduction on the accuracy of machine learning. 2019 9th Annual Information Technology, Electromechanical Engineering and Microelectronics Conference (IEMECON); March 2019; Jaipur, India. pp. 279–283. - DOI
-
- Xu X., Liang T., Zhu J., Zheng D., Sun T. Review of classical dimensionality reduction and sample selection methods for large-scale data processing. Neurocomputing . 2019;328:5–15. doi: 10.1016/j.neucom.2018.02.100. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

