Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics
- PMID: 34824791
- PMCID: PMC8601879
- DOI: 10.1002/ece3.8250
Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics
Abstract
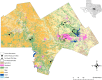
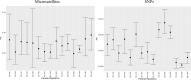
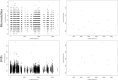
Comparisons of microsatellites and single-nucleotide polymorphisms (SNPs) have found that SNPs outperform microsatellites in population genetic analyses, questioning the continued utility of microsatellites in population and landscape genetics. Yet, highly polymorphic markers may be of value in species that have reduced genetic variation. This study repeated previous analyses that used microsatellites with SNPs developed from ddRAD sequencing in the black-capped vireo source-sink system. SNPs provided greater resolution of genetic diversity, population differentiation, and migrant detection but could not reconstruct parentage relationships due to insufficient heterozygosities. The biological inferences made by both sets of markers were similar: asymmetrical gene flow from source sites to the remaining sink sites. With the landscape genetic analyses, we found different results between the two molecular markers, but associations of the top environmental features (riparian, open habitat, agriculture, and human development) with dispersal estimates were shared between marker types. Despite the higher precision of SNPs, we find that microsatellites effectively uncover population processes and patterns and are superior for parentage analyses in this species with reduced genetic diversity. This study illustrates the continued applicability and relevance of microsatellites in population genetic research.
Keywords: genomics; molecular markers; next‐generation sequencing; parentage; population structure; resistance surfaces.
© 2021 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to report.
Figures

References
-
- Adamack, A. T. , & Gruber, B. (2014). PopGenReport: Simplifying basic population genetic analyses in R. Methods in Ecology and Evolution, 5(4), 384–387.
-
- Altman, N. , & Krzywinski, M. (2016). Points of significance: P values and the search for significance. Nature Methods, 14(1), 3–4. 10.1038/nmeth.4120 - DOI
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

