Identification of Candidate Genes for Drought Tolerance at Maize Seedlings Using Genome-Wide Association
- PMID: 34825009
- PMCID: PMC8590722
- DOI: 10.30498/ijb.2021.209324.2637
Identification of Candidate Genes for Drought Tolerance at Maize Seedlings Using Genome-Wide Association
Abstract
Background: Drought stress is a serious threat that limit maize growth and production.
Objectives: The assessment tolerance level of maize by measuring changes in the main biochemical and physiological indicators under drought stress.
Material and methods: We performed a genome-wide association analysis of biochemical and physiological indicators using an elite association panel.
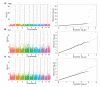
Results: The results revealed that eight significant SNPs (p<0.05/N) located in eight genes that are distributed on different chromosomes were associated with drought resistance indices under drought stress. Among these genes, four genes were linked via the associated SNPs with drought-resistance indices of the malondialdehyde activity (MDA), three genes were linked with drought resistance indexes of the superoxide dismutase activity (SOD), and one gene was linked with drought resistance indexes of relative conductivity (REC). The candidate genes functioned as transcription factors, enzymes, and transporters, which included trehalase, the AP2/EREB160 transcription factor, and glutathione S-transferase and also encoded a gene of unknown function. These genes may be directly or indirectly involved in drought resistance. The expression levels of ZmEREB160 responded to ABA and drought stress.
Conclusions: These results provided good information to understand the genetic basis of variation in drought resistance indices of biochemical and physiological indicators during drought stress.
Keywords: Drought-resistance indices; Genome-wide association study; Maize; Physiological and biochemical traits.
Copyright: © 2021 The Author(s); Published by Iranian Journal of Biotechnology.
Figures


References
-
- Zhang T, Yu LX, Zheng P, Li Y, Rivera M, Main D, Greene SL. Identification of Loci Associated with Drought Resistance Traits in Heterozygous Autotetraploid Alfalfa (Medicago sativa L.) Using Genome-Wide Association Studies with Genotyping by Sequencing. PLoS One. 2015;10(9):e0138931. doi: 10.1371/journal.pone.0138931. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
