Improved epicardial cardiac fibroblast generation from iPSCs
- PMID: 34826415
- PMCID: PMC9048147
- DOI: 10.1016/j.yjmcc.2021.11.011
Improved epicardial cardiac fibroblast generation from iPSCs
Abstract
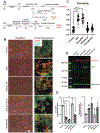
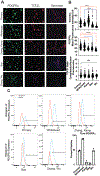
Since the initial isolation of human embryonic stem cells and subsequent discovery of reprogramming methods for somatic cells, thousands of protocols have been developed to create each of the hundreds of cell types found in-vivo with significant focus on disease-prone systems, e.g., cardiovascular. Robust protocols exist for many of these cell types, except for cardiac fibroblasts (CF). Very recently, several competing methods have been developed to generate these cells through a developmentally conserved epicardial pathway. Such methods generate epicardial cells, but here we report that prolonged exposure to growth factors such as bFGF induces fibroblast spindle-like morphology and similar chromatin architecture to primary CFs. Media conditions for growth and assays are provided, as well as suggestions for seeding densities and timepoints for protein harvest of extracellular matrix. We demonstrate marker expression and matrix competency of resultant cells as shown next to primary human cardiac fibroblasts. These methods provide additional guidance to the original protocol and result in an increasingly stable phenotype.
Keywords: ATAC-sequencing; Cardiac fibroblast; Differentiation; Epicardial; iPSC.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

