CRISPR Element Patterns vs. Pathoadaptability of Clinical Pseudomonas aeruginosa Isolates from a Medical Center in Moscow, Russia
- PMID: 34827239
- PMCID: PMC8615150
- DOI: 10.3390/antibiotics10111301
CRISPR Element Patterns vs. Pathoadaptability of Clinical Pseudomonas aeruginosa Isolates from a Medical Center in Moscow, Russia
Abstract
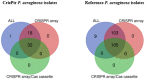
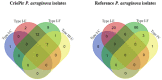
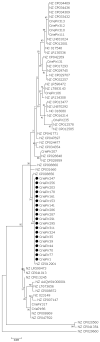
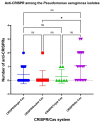
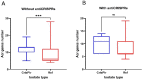
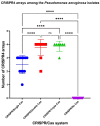
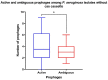
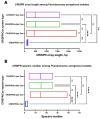
Pseudomonas aeruginosa is a member of the ESKAPE opportunistic pathogen group, which includes six species of the most dangerous microbes. This pathogen is characterized by the rapid acquisition of antimicrobial resistance, thus causing major healthcare concerns. This study presents a comprehensive analysis of clinical P. aeruginosa isolates based on whole-genome sequencing data. The isolate collection studied was characterized by a variety of clonal lineages with a domination of high-risk epidemic clones and different CRISPR/Cas element patterns. This is the first report on the coexistence of two and even three different types of CRISPR/Cas systems simultaneously in Russian clinical strains of P. aeruginosa. The data include molecular typing and genotypic antibiotic resistance determination, as well as the phylogenetic analysis of the full-length cas gene and anti-CRISPR genes sequences, predicted prophage sequences, and conducted a detailed CRISPR array analysis. The differences between the isolates carrying different types and quantities of CRISPR/Cas systems were investigated. The pattern of virulence factors in P. aeruginosa isolates lacking putative CRISPR/Cas systems significantly differed from that of samples with single or multiple putative CRISPR/Cas systems. We found significant correlations between the numbers of prophage sequences, antibiotic resistance genes, and virulence genes in P. aeruginosa isolates with different patterns of CRISPR/Cas-elements. We believe that the data presented will contribute to further investigations in the field of bacterial pathoadaptability, including antimicrobial resistance and the role of CRISPR/Cas systems in the plasticity of the P. aeruginosa genome.
Keywords: Pseudomonas aeruginosa; WGS; antibiotic resistance; multiple CRISPR/Cas systems; pathoadaptability; virulence factors.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- WHO . Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities. World Health Organization; Geneva, Switzerland: 2017. - PubMed
-
- Edelstein M.V., Skleenova E.N., Shevchenko O.V., D’Souza J.W., Tapalski D.V., Azizov I.S., Sukhorukova M.V., Pavlukov R.A., Kozlov R.S., Toleman M.A., et al. Spread of extensively resistant VIM-2-positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: A longitudinal epidemiological and clinical study. Lancet Infect. Dis. 2013;13:867–876. doi: 10.1016/S1473-3099(13)70168-3. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

