Investigating the Reciprocal Interrelationships among the Ruminal Microbiota, Metabolome, and Mastitis in Early Lactating Holstein Dairy Cows
- PMID: 34827839
- PMCID: PMC8614428
- DOI: 10.3390/ani11113108
Investigating the Reciprocal Interrelationships among the Ruminal Microbiota, Metabolome, and Mastitis in Early Lactating Holstein Dairy Cows
Abstract
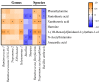
Mastitis in dairy cow significantly affects animal performance, ultimately reducing profitability. The reciprocal interrelationships among ruminal microbiota, metabolome, and mastitis combining early inflammatory factors (serum proinflammatory cytokines) in lactating dairy cows has not been explored, thus, this study evaluated these reciprocal interrelationships in early lactating Holstein dairy cows to identify potential microbial biomarkers and their relationship with ruminal metabolites. The ruminal fluid was sampled from 8 healthy and 8 mastitis cows for the microbiota and metabolite analyses. The critical ruminal microbial biomarkers and metabolites related to somatic cell counts (SCC) and serum proinflammatory cytokines were identified by the linear discriminant analysis effect size (LEfSe) algorithm and Spearman's correlation analysis, respectively. The SCC level and proinflammatory cytokines positively correlated with Sharpea and negatively correlated with Ruminococcaceae UCG-014, Ruminococcus flavefaciens, and Treponema saccharophilum. Furthermore, the metabolites xanthurenic acid, and 1-(1H-benzo[d]imidazol-2-yl) ethan-1-ol positively correlated with microbial biomarkers of healthy cows, whereas, xanthine, pantothenic acid, and anacardic acid were negatively correlated with the microbial biomarkers of mastitis cows. In conclusion, Ruminococcus flavefaciens and Treponema saccharophilum are potential strains for improving the health of dairy cows. The current study provides a novel perspective to assist in targeting the ruminal microbiota with preventive/therapeutic strategies against inflammatory diseases in the future.
Keywords: Holstein dairy cows; mastitis; metabolome; ruminal microbiota.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Barbosa L.F.S.P., Oliveira W.V.C., Pereira M.H.C., Moreira M.B., Vasconcelos C.G.C., Silper B.F., Cerri R.L.A., Vasconcelos J.L.M. Somatic cell count and type of intramammary infection impacts fertility from in vitro produced embryo transfer. Theriogenology. 2018;108:291–296. doi: 10.1016/j.theriogenology.2017.12.025. - DOI - PubMed
-
- Ribeiro E.S., Gomes G., Greco L.F., Cerri R.L.A., Monteiro P.L.J., Jr., Lima F.S., Bisinotto R.S., Thatcher W.W., Santos J.E.P. Carryover effect of postpartum inflam-matory diseases on developmental biology and fertility in lactating dairy cows. J. Dairy Sci. 2016;99:2201–2222. doi: 10.3168/jds.2015-10337. - DOI - PubMed
-
- Falentin H., Rault L., Nicolas A., Bouchard D.S., Lassalas J., Lamberton P., Aubry J.M., Marnet P.G., Loir Y.L., Even S. Bovine teat microbiome analysis revealed reduced alpha diversity and significant changes in taxonomic profiles in quarters with a history of mastitis. Front. Microbiol. 2016;7:480. doi: 10.3389/fmicb.2016.00480. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

