NOV/CCN3 Promotes Cell Migration and Invasion in Intrahepatic Cholangiocarcinoma via miR-92a-3p
- PMID: 34828265
- PMCID: PMC8621878
- DOI: 10.3390/genes12111659
NOV/CCN3 Promotes Cell Migration and Invasion in Intrahepatic Cholangiocarcinoma via miR-92a-3p
Abstract
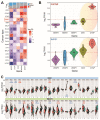
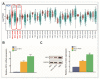
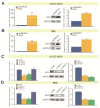
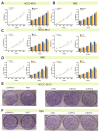
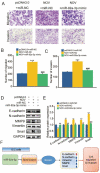
Intrahepatic cholangiocarcinoma (ICC) is a common type of human cancer with a poor prognosis, and investigating the potential molecular mechanisms that can contribute to gene diagnosis and therapy. Herein, based on the recently concerned vertebrate-specific Cyr61/CTGF/NOV (CCN) gene family because of its important roles in diverse diseases, we obtained NOV/CCN3 to query for its potential roles in tumorigenesis via bioinformatics analysis. Experimental validations confirmed that both NOV mRNA and protein are up-regulated in two ICC cell lines, suggesting that it may promote cell migration and invasion by promoting EMT. To elucidate the detailed regulatory mechanism, miR-92a-3p is screened and identified as a negative regulatory small RNA targeting NOV, and further experimental validation demonstrates that miR-92a-3p contributes to NOV-mediated migration and invasion of ICC via the Notch signaling pathway. Our study reveals that NOV may be a potential target for diagnosing and treating ICC, which will provide experimental data and molecular theoretical foundation for cancer treatment, particularly for future precision medicine.
Keywords: NOV/CCN3; cell migration; intrahepatic cholangiocarcinoma; invasion; miR-92a-3p.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Yin Yang 1-induced LINC00667 up-regulates pyruvate dehydrogenase kinase 1 to promote proliferation, migration and invasion of cholangiocarcinoma cells by sponging miR-200c-3p.Hum Cell. 2021 Jan;34(1):187-200. doi: 10.1007/s13577-020-00448-1. Epub 2020 Oct 10. Hum Cell. 2021. PMID: 33040228
-
miR-204 inhibits epithelial to mesenchymal transition by targeting slug in intrahepatic cholangiocarcinoma cells.Cell Physiol Biochem. 2013;32(5):1331-41. doi: 10.1159/000354531. Epub 2013 Nov 22. Cell Physiol Biochem. 2013. PMID: 24280681
-
LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma Through Suppressing miR-152.Dig Dis Sci. 2017 Nov;62(11):3050-3058. doi: 10.1007/s10620-017-4759-8. Epub 2017 Sep 18. Dig Dis Sci. 2017. PMID: 28921383
-
The role of microRNAs in intrahepatic cholangiocarcinoma.J Cell Mol Med. 2017 Jan;21(1):177-184. doi: 10.1111/jcmm.12951. Epub 2016 Sep 13. J Cell Mol Med. 2017. PMID: 27619971 Free PMC article. Review.
-
The Role of microRNAs in Cholangiocarcinoma.Int J Mol Sci. 2021 Jul 16;22(14):7627. doi: 10.3390/ijms22147627. Int J Mol Sci. 2021. PMID: 34299246 Free PMC article. Review.
Cited by
-
CCN3/NOV promotes metastasis and tumor progression via GPNMB-induced EGFR activation in triple-negative breast cancer.Cell Death Dis. 2023 Feb 3;14(2):81. doi: 10.1038/s41419-023-05608-3. Cell Death Dis. 2023. PMID: 36737605 Free PMC article.
-
A comprehensive multi-omics analysis reveals molecular features associated with cancer via RNA cross-talks in the Notch signaling pathway.Comput Struct Biotechnol J. 2022 Jul 26;20:3972-3985. doi: 10.1016/j.csbj.2022.07.036. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35950189 Free PMC article.
-
Distinct cholangiocarcinoma cell migration in 2D monolayer and 3D spheroid culture based on galectin-3 expression and localization.Front Oncol. 2023 Jan 12;12:999158. doi: 10.3389/fonc.2022.999158. eCollection 2022. Front Oncol. 2023. PMID: 36713574 Free PMC article.
-
Decidualization of human endometrial stromal cells requires steroid receptor coactivator-3.Front Reprod Health. 2022 Nov 24;4:1033581. doi: 10.3389/frph.2022.1033581. eCollection 2022. Front Reprod Health. 2022. PMID: 36505394 Free PMC article.
-
Activating mutations remodel the chromatin accessibility landscape to drive distinct regulatory networks in KMT2A-rearranged acute leukemia.Hemasphere. 2024 Sep 26;8(9):e70006. doi: 10.1002/hem3.70006. eCollection 2024 Sep. Hemasphere. 2024. PMID: 39329074 Free PMC article.
References
-
- Chinchilla-López P., Aguilar-Olivos N.E., García-Gómez J., Hernández-Alejandro K.K., Chablé-Montero F., Motola-Kuba D., Patel T., Méndez-Sánchez N. Prevalence, Risk Factors, and Survival of Patients with Intrahepatic Cholangiocarcinoma. Ann. Hepatol. 2017;16:565–568. doi: 10.5604/01.3001.0010.0293. - DOI - PubMed
-
- Czito B.G., Anscher M.S., Willett C.G. Radiation therapy in the treatment of cholangiocarcinoma. Oncology. 2006;20:873–884. - PubMed
-
- Endo I., Gonen M., Yopp A.C., Dalal K.M., Zhou Q., Klimstra D., D’Angelica M., DeMatteo R.P., Fong Y., Schwartz L., et al. Intrahepatic cholangiocarcinoma: Rising frequency, improved survival, and determinants of outcome after resection. Ann. Surg. 2008;248:84–96. doi: 10.1097/SLA.0b013e318176c4d3. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

