Molecular Classification and Interpretation of Amyotrophic Lateral Sclerosis Using Deep Convolution Neural Networks and Shapley Values
- PMID: 34828360
- PMCID: PMC8626003
- DOI: 10.3390/genes12111754
Molecular Classification and Interpretation of Amyotrophic Lateral Sclerosis Using Deep Convolution Neural Networks and Shapley Values
Abstract
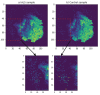
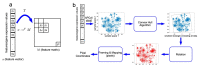
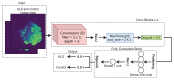
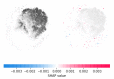
Amyotrophic lateral sclerosis (ALS) is a prototypical neurodegenerative disease characterized by progressive degeneration of motor neurons to severely effect the functionality to control voluntary muscle movement. Most of the non-additive genetic aberrations responsible for ALS make its molecular classification very challenging along with limited sample size, curse of dimensionality, class imbalance and noise in the data. Deep learning methods have been successful in many other related areas but have low minority class accuracy and suffer from the lack of explainability when used directly with RNA expression features for ALS molecular classification. In this paper, we propose a deep-learning-based molecular ALS classification and interpretation framework. Our framework is based on training a convolution neural network (CNN) on images obtained from converting RNA expression values into pixels based on DeepInsight similarity technique. Then, we employed Shapley additive explanations (SHAP) to extract pixels with higher relevance to ALS classifications. These pixels were mapped back to the genes which made them up. This enabled us to classify ALS samples with high accuracy for a minority class along with identifying genes that might be playing an important role in ALS molecular classifications. Taken together with RNA expression images classified with CNN, our preliminary analysis of the genes identified by SHAP interpretation demonstrate the value of utilizing Machine Learning to perform molecular classification of ALS and uncover disease-associated genes.
Keywords: ALS; classification; interpretation; machine learning; target identification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Yin B., Balvert M., van der Spek R.A., Dutilh B.E., Bohté S., Veldink J., Schönhuth A. Using the structure of genome data in the design of deep neural networks for predicting amyotrophic lateral sclerosis from genotype. Bioinformatics. 2019;35:i538–i547. doi: 10.1093/bioinformatics/btz369. - DOI - PMC - PubMed
-
- Amyotrophic Lateral Sclerosis (ALS) Fact Sheet | National Institute of Neurological Disorders and Stroke. [(accessed on 15 February 2021)]; Available online: https://www.ninds.nih.gov/Disorders/Patient-Caregiver-Education/Fact-She....
-
- Van Rheenen W., Shatunov A., Dekker A.M., McLaughlin R.L., Diekstra F.P., Pulit S.L., Van Der Spek R.A., Võsa U., De Jong S., Robinson M.R., et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 2016;48:1043–1048. doi: 10.1038/ng.3622. - DOI - PMC - PubMed
-
- Arloth J., Eraslan G., Andlauer T.F., Martins J., Iurato S., Kühnel B., Waldenberger M., Frank J., Gold R., Hemmer B., et al. DeepWAS: Multivariate genotype-phenotype associations by directly integrating regulatory information using deep learning. PLoS Comput. Biol. 2020;16:e1007616. doi: 10.1371/journal.pcbi.1007616. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

