Sex Chromosomes and Master Sex-Determining Genes in Turtles and Other Reptiles
- PMID: 34828428
- PMCID: PMC8622242
- DOI: 10.3390/genes12111822
Sex Chromosomes and Master Sex-Determining Genes in Turtles and Other Reptiles
Abstract
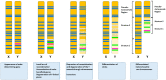
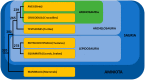
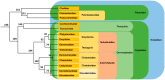
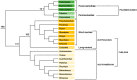
Among tetrapods, the well differentiated heteromorphic sex chromosomes of birds and mammals have been highly investigated and their master sex-determining (MSD) gene, Dmrt1 and SRY, respectively, have been identified. The homomorphic sex chromosomes of reptiles have been the least studied, but the gap with birds and mammals has begun to fill. This review describes our current knowledge of reptilian sex chromosomes at the cytogenetic and molecular level. Most of it arose recently from various studies comparing male to female gene content. This includes restriction site-associated DNA sequencing (RAD-Seq) experiments in several male and female samples, RNA sequencing and identification of Z- or X-linked genes by male/female comparative transcriptome coverage, and male/female transcriptomic or transcriptome/genome substraction approaches allowing the identification of Y- or W-linked transcripts. A few putative master sex-determining (MSD) genes have been proposed, but none has been demonstrated yet. Lastly, future directions in the field of reptilian sex chromosomes and their MSD gene studies are considered.
Keywords: Genetic Sex Determination (GSD); RAD-seq; genome coverage; homologous genes; lizards; snakes; squamates.
Conflict of interest statement
The author declares no conflict of interest.
Figures




References
-
- Imarazene B., Du K., Beille S., Jouanno E., Feron R., Pan Q., Torres-Paz J., Lopez-Roques C., Castinel A., Gil L., et al. A supernumerary “B-sex” chromosome drives male sex determination in the Pachón cavefish, Astyanax mexicanus. Curr. Biol. 2021;31:4800–4809. doi: 10.1016/j.cub.2021.08.030. - DOI - PMC - PubMed
-
- Sinclair A.H., Berta P., Palmer M.S., Hawkins J.R., Griffiths B.L., Smith M.J., Foster J.W., Frischauf A.M., Lovell-Badge R., Goodfellow P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

