Tracking SARS-CoV-2: Novel Trends and Diagnostic Strategies
- PMID: 34829328
- PMCID: PMC8621220
- DOI: 10.3390/diagnostics11111981
Tracking SARS-CoV-2: Novel Trends and Diagnostic Strategies
Abstract
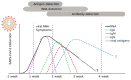
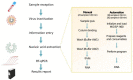
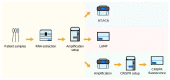
The COVID-19 pandemic has had an enormous impact on economies and health systems globally, therefore a top priority is the development of increasingly better diagnostic and surveillance alternatives to slow down the spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In order to establish massive testing and contact tracing policies, it is crucial to have a clear view of the diagnostic options available and their principal advantages and drawbacks. Although classical molecular methods such as RT-qPCR are broadly used, diagnostic alternatives based on technologies such as LAMP, antigen, serological testing, or the application of novel technologies such as CRISPR-Cas for diagnostics, are also discussed. The present review also discusses the most important automation strategies employed to increase testing capability. Several serological-based diagnostic kits are presented, as well as novel nanotechnology-based diagnostic methods. In summary, this review provides a clear diagnostic landscape of the most relevant tools to track COVID-19.
Keywords: COVID-19; CRISPR-based diagnostics; antigen testing; automation; nanotechnology-based diagnostics; nucleic acid amplification test.
Conflict of interest statement
The authors declare having no competing or financial interests or personal relationships related to this manuscript.
Figures



References
-
- World Health Organization . WHO Consensus Document on Theepidemiology of Severe Acuterespiratory Syndrome (SARS) Department of Communicable Disease Surveillance and Response; Geneva, Switzerland: 2003.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

