The Role of NKL Homeobox Genes in T-Cell Malignancies
- PMID: 34829904
- PMCID: PMC8615965
- DOI: 10.3390/biomedicines9111676
The Role of NKL Homeobox Genes in T-Cell Malignancies
Abstract
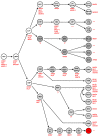
Homeobox genes encode transcription factors controlling basic developmental processes. The homeodomain is encoded by the homeobox and mediates sequence-specific DNA binding and interaction with cofactors, thus operating as a basic regulatory platform. Similarities in their homeobox sequences serve to arrange these genes in classes and subclasses, including NKL homeobox genes. In accordance with their normal functions, deregulated homeobox genes contribute to carcinogenesis along with hematopoietic malignancies. We have recently described the physiological expression of eleven NKL homeobox genes in the course of hematopoiesis and termed this gene expression pattern NKL-code. Due to the developmental impact of NKL homeobox genes these data suggest a key role for their activity in the normal regulation of hematopoietic cell differentiation including T-cells. On the other hand, aberrant overexpression of NKL-code members or ectopical activation of non-code members has been frequently reported in lymphoid and myeloid leukemia/lymphoma, demonstrating their oncogenic impact in the hematopoietic compartment. Here, we provide an overview of the NKL-code in normal hematopoiesis and discuss the oncogenic role of deregulated NKL homeobox genes in T-cell malignancies.
Keywords: NKL-code; T-all; homeobox; homeodomain; lymphoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
NKL-Code in Normal and Aberrant Hematopoiesis.Cancers (Basel). 2021 Apr 19;13(8):1961. doi: 10.3390/cancers13081961. Cancers (Basel). 2021. PMID: 33921702 Free PMC article. Review.
-
Deregulated NKL Homeobox Genes in B-Cell Lymphoma.Cancers (Basel). 2019 Nov 26;11(12):1874. doi: 10.3390/cancers11121874. Cancers (Basel). 2019. PMID: 31779217 Free PMC article. Review.
-
NKL Homeobox Genes NKX2-3 and NKX2-4 Deregulate Megakaryocytic-Erythroid Cell Differentiation in AML.Int J Mol Sci. 2021 Oct 22;22(21):11434. doi: 10.3390/ijms222111434. Int J Mol Sci. 2021. PMID: 34768865 Free PMC article.
-
NKL homeobox gene activities in B-cell development and lymphomas.PLoS One. 2018 Oct 11;13(10):e0205537. doi: 10.1371/journal.pone.0205537. eCollection 2018. PLoS One. 2018. PMID: 30308041 Free PMC article.
-
NKL homeobox gene activities in normal and malignant myeloid cells.PLoS One. 2019 Dec 11;14(12):e0226212. doi: 10.1371/journal.pone.0226212. eCollection 2019. PLoS One. 2019. PMID: 31825998 Free PMC article.
Cited by
-
Establishment of the Myeloid TBX-Code Reveals Aberrant Expression of T-Box Gene TBX1 in Chronic Myeloid Leukemia.Int J Mol Sci. 2023 Dec 19;25(1):32. doi: 10.3390/ijms25010032. Int J Mol Sci. 2023. PMID: 38203204 Free PMC article.
References
-
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

