Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells
- PMID: 34829947
- PMCID: PMC8615743
- DOI: 10.3390/biomedicines9111718
Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells
Abstract
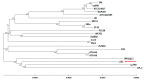
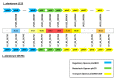
Lactiplantibacillus plantarum is a diverse species that includes nomadic strains isolated from a variety of environmental niches. Several L. plantarum strains are being incorporated in fermented foodstuffs as starter cultures, while some of them have also been characterized as probiotics. In this study, we present the draft genome sequence of L. plantarum L125, a potential probiotic strain presenting biotechnological interest, originally isolated from a traditional fermented meat product. Phylogenetic and comparative genomic analysis with other potential probiotic L. plantarum strains were performed to determine its evolutionary relationships. Furthermore, we located genes involved in the probiotic phenotype by whole genome annotation. Indeed, genes coding for proteins mediating host-microbe interactions and bile salt, heat and cold stress tolerance were identified. Concerning the potential health-promoting attributes of the novel strain, we determined that L. plantarum L125 carries an incomplete plantaricin gene cluster, in agreement with previous in vitro findings, where no bacteriocin-like activity was detected. Moreover, we showed that cell-free culture supernatant (CFCS) of L. plantarum L125 exerts anti-proliferative, anti-clonogenic and anti-migration activity against the human colon adenocarcinoma cell line, HT-29. Conclusively, L. plantarum L125 presents desirable probiotic traits. Future studies will elucidate further its biological and health-related properties.
Keywords: Lactiplantibacillus plantarum; anti-proliferative activity; comparative genomics; genomics; phylogenetic analysis; probiotics; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures





Similar articles
-
Whole-Genome Sequencing, Phylogenetic and Genomic Analysis of Lactiplantibacillus pentosus L33, a Potential Probiotic Strain Isolated From Fermented Sausages.Front Microbiol. 2021 Oct 26;12:746659. doi: 10.3389/fmicb.2021.746659. eCollection 2021. Front Microbiol. 2021. PMID: 34764945 Free PMC article.
-
Probiotic and anti-inflammatory properties of Lactiplantibacillus plantarum MKTJ24 isolated from an artisanal fermented fish of North-east India.N Biotechnol. 2024 Nov 25;83:121-132. doi: 10.1016/j.nbt.2024.07.005. Epub 2024 Aug 6. N Biotechnol. 2024. PMID: 39111568
-
Genomic Characterization of Lactiplantibacillus plantarum Strains: Potential Probiotics from Ethiopian Traditional Fermented Cottage Cheese.Genes (Basel). 2024 Oct 29;15(11):1389. doi: 10.3390/genes15111389. Genes (Basel). 2024. PMID: 39596588 Free PMC article.
-
Bioprospecting Antimicrobials from Lactiplantibacillus plantarum: Key Factors Underlying Its Probiotic Action.Int J Mol Sci. 2021 Nov 8;22(21):12076. doi: 10.3390/ijms222112076. Int J Mol Sci. 2021. PMID: 34769500 Free PMC article. Review.
-
Health-Promoting Role of Lactiplantibacillus plantarum Isolated from Fermented Foods.Microorganisms. 2021 Feb 10;9(2):349. doi: 10.3390/microorganisms9020349. Microorganisms. 2021. PMID: 33578806 Free PMC article. Review.
Cited by
-
Probiotic Potential and Safety Assessment of Lactiplantibacillus plantarum cqf-43 and Whole-Genome Sequence Analysis.Int J Mol Sci. 2023 Dec 17;24(24):17570. doi: 10.3390/ijms242417570. Int J Mol Sci. 2023. PMID: 38139398 Free PMC article.
-
The Gut Microbiota and Colorectal Cancer: Understanding the Link and Exploring Therapeutic Interventions.Biology (Basel). 2025 Feb 28;14(3):251. doi: 10.3390/biology14030251. Biology (Basel). 2025. PMID: 40136508 Free PMC article. Review.
-
The Impacts of Lactiplantibacillus plantarum on the Functional Properties of Fermented Foods: A Review of Current Knowledge.Microorganisms. 2022 Apr 15;10(4):826. doi: 10.3390/microorganisms10040826. Microorganisms. 2022. PMID: 35456875 Free PMC article. Review.
-
Comparative genomics of food-derived probiotic Lactiplantibacillus plantarum K25 reveals its hidden potential, compactness, and efficiency.Front Microbiol. 2023 Jun 14;14:1214478. doi: 10.3389/fmicb.2023.1214478. eCollection 2023. Front Microbiol. 2023. PMID: 37455721 Free PMC article.
-
Probiotic-Derived Bioactive Compounds in Colorectal Cancer Treatment.Microorganisms. 2023 Jul 27;11(8):1898. doi: 10.3390/microorganisms11081898. Microorganisms. 2023. PMID: 37630458 Free PMC article. Review.
References
-
- Siezen R.J., Tzeneva V.A., Castioni A., Wels M., Phan H.T., Rademaker J.L., Starrenburg M.J., Kleerebezem M., Molenaar D., van Hylckama Vlieg J.E. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010;12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. - DOI - PubMed
-
- Zheng J., Wittouck S., Salvetti E., Franz C., Harris H.M.B., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. - DOI - PubMed
-
- Martino M.E., Bayjanov J.R., Caffrey B.E., Wels M., Joncour P., Hughes S., Gillet B., Kleerebezem M., van Hijum S.A., Leulier F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016;18:4974–4989. doi: 10.1111/1462-2920.13455. - DOI - PubMed
-
- Ruiz M.J., Zbrun M.V., Signorini M.L., Zimmermann J.A., Soto L.P., Rosmini M.R., Frizzo L.S. In vitro screening and in vivo colonization pilot model of Lactobacillus plantarum LP5 and Campylobacter coli DSPV 458 in mice. Arch. Microbiol. 2021;203:4161–4171. doi: 10.1007/s00203-021-02385-5. - DOI - PubMed
Grants and funding
- 5047285/European Regional Development Fund (ERDF) and national resources
- 5002780/Operational Program Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020) and co-financed by Greece and the European Union
- 5046239/Operational Program Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020) and co-financed by Greece and the European Union
- 5047291/Operational Program Competitiveness, Entrepreneurship and Innovation (NSRF 2014-2020) and co-financed by Greece and the European Union
- 5056124/European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

