Xanthohumol Is a Potent Pan-Inhibitor of Coronaviruses Targeting Main Protease
- PMID: 34830015
- PMCID: PMC8624673
- DOI: 10.3390/ijms222212134
Xanthohumol Is a Potent Pan-Inhibitor of Coronaviruses Targeting Main Protease
Abstract
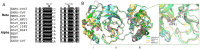
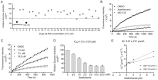
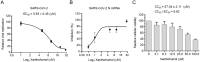
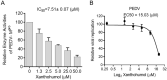
Coronaviruses cause diseases in humans and livestock. The SARS-CoV-2 is infecting millions of human beings, with high morbidity and mortality worldwide. The main protease (Mpro) of coronavirus plays a pivotal role in viral replication and transcription, which, in theory, is an attractive drug target for antiviral drug development. It has been extensively discussed whether Xanthohumol is able to help COVID-19 patients. Here, we report that Xanthohumol, a small molecule in clinical trials from hops (Humulus lupulus), was a potent pan-inhibitor for various coronaviruses by targeting Mpro, for example, betacoronavirus SARS-CoV-2 (IC50 value of 1.53 μM), and alphacoronavirus PEDV (IC50 value of 7.51 μM). Xanthohumol inhibited Mpro activities in the enzymatical assays, while pretreatment with Xanthohumol restricted the SARS-CoV-2 and PEDV replication in Vero-E6 cells. Therefore, Xanthohumol is a potent pan-inhibitor of coronaviruses and an excellent lead compound for further drug development.
Keywords: PEDV; SARS-CoV-2; Xanthohumol; coronavirus; natural product.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Edwards C.E., Yount B.L., Graham R.L., Leist S.R., Hou Y.J., Dinnon K.H., Sims A.C., Swanstrom J., Gully K., Scobey T.D., et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl. Acad. Sci. USA. 2020;117:26915–26925. doi: 10.1073/pnas.2001046117. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

