There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages
- PMID: 34830050
- PMCID: PMC8620503
- DOI: 10.3390/ijms222212172
There Is Strength in Numbers: Quantitation of Fc Gamma Receptors on Murine Tissue-Resident Macrophages
Abstract
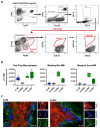
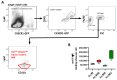
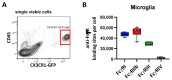
Many of the effector functions of antibodies rely on the binding of antibodies/immune complexes to cellular Fcγ receptors (FcγRs). Since the majority of innate immune effector cells express both activating and inhibitory Fc receptors, the outcome of the binding of immune complexes to cells of a given population is influenced by the relative affinities of the respective IgG subclasses to these receptors, as well as by the numbers of activating and inhibitory FcγRs on the cell surface. A group of immune cells that has come into focus more recently is the various subsets of tissue-resident macrophages. The central functions of FcγRs on tissue macrophages include the clearance of opsonized pathogens, the removal of small immune complexes from the circulation and the depletion of antibody-opsonized cells in the therapy of autoimmunity and cancer. Despite these essential functions of FcγRs on tissue-resident macrophages, an in-depth quantification of FcγRs is lacking. Thus, the aim of our current study was to quantify the various Fcγ receptors on macrophages in murine liver, lung, kidney, brain, skin and spleen. Our study identified a pronounced heterogeneity between FcγR expression patterns of the different tissue macrophages, which may reflect their specialized functions within their unique niches in different organ environments.
Keywords: Fc receptors; Kupffer cells; Langerhans cells; alveolar macrophages; antibodies; dermal macrophages; interstitial macrophages; kidney resident macrophages; macrophages; microglia; splenic macrophages.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

