Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF
- PMID: 34830080
- PMCID: PMC8621504
- DOI: 10.3390/ijms222212199
Ets1 Promotes the Differentiation of Post-Selected iNKT Cells through Regulation of the Expression of Vα14Jα18 T Cell Receptor and PLZF
Abstract
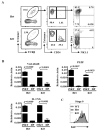
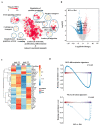
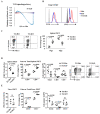
The transcription factor Ets1 is essential for the development/differentiation of invariant Natural Killer T (iNKT) cells at multiple stages. However, its mechanisms of action and target genes in iNKT cells are still elusive. Here, we show that Ets1 is required for the optimal expression of the Vα14Jα18 T cell receptor (TCR) in post-selected thymic iNKT cells and their immediate differentiation. Ets1 is also critical for maintaining the peripheral homeostasis of iNKT cells, which is a role independent of the expression of the Vα14Jα18 TCR. Genome-wide transcriptomic analyses of post-selected iNKT cells further reveal that Ets1 controls leukocytes activation, proliferation differentiation, and leukocyte-mediated immunity. In addition, Ets1 regulates the expression of ICOS and PLZF in iNKT cells. More importantly, restoring the expression of PLZF and the Vα14Jα18 TCR partially rescues the differentiation of iNKT cells in the absence of Ets1. Taken together, our results establish a detailed molecular picture of how Ets1 regulates the stepwise differentiation of iNKT cells.
Keywords: Ets1; ICOS; PLZF; iNKT cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Monoclonal Invariant NKT (iNKT) Cell Mice Reveal a Role for Both Tissue of Origin and the TCR in Development of iNKT Functional Subsets.J Immunol. 2017 Jul 1;199(1):159-171. doi: 10.4049/jimmunol.1700214. Epub 2017 Jun 2. J Immunol. 2017. PMID: 28576977 Free PMC article.
-
Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc.Proc Natl Acad Sci U S A. 2009 May 26;106(21):8641-6. doi: 10.1073/pnas.0812255106. Epub 2009 May 7. Proc Natl Acad Sci U S A. 2009. PMID: 19423665 Free PMC article.
-
Quantitative Difference in PLZF Protein Expression Determines iNKT Lineage Fate and Controls Innate CD8 T Cell Generation.Cell Rep. 2019 May 28;27(9):2548-2557.e4. doi: 10.1016/j.celrep.2019.05.012. Cell Rep. 2019. PMID: 31141681 Free PMC article.
-
The Role of Autophagy in iNKT Cell Development.Front Immunol. 2018 Nov 14;9:2653. doi: 10.3389/fimmu.2018.02653. eCollection 2018. Front Immunol. 2018. PMID: 30487800 Free PMC article. Review.
-
The ins and outs of type I iNKT cell development.Mol Immunol. 2019 Jan;105:116-130. doi: 10.1016/j.molimm.2018.09.023. Epub 2018 Nov 28. Mol Immunol. 2019. PMID: 30502719 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- 107-2320-B-002-027-MY3 to YTC, 106-2311-B-650-001 and 107-2311-B-650-001 to TST/Ministry of Science and Technology
- CMRPG3L0191 to TST/Linkou Chang Gung Memorial Hospital
- W81XWH-11-1-0492/United States Department of Defense
- HHSN272201300006C/AI/NIAID NIH HHS/United States
- 110-S4897 and 110-EDN10/National Taiwan University
LinkOut - more resources
Full Text Sources
Miscellaneous

