Identification of Potential Long Non-Coding RNA Candidates that Contribute to Triple-Negative Breast Cancer in Humans through Computational Approach
- PMID: 34830241
- PMCID: PMC8619140
- DOI: 10.3390/ijms222212359
Identification of Potential Long Non-Coding RNA Candidates that Contribute to Triple-Negative Breast Cancer in Humans through Computational Approach
Abstract
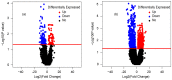
Breast cancer (BC) is the most frequent malignancy identified in adult females, resulting in enormous financial losses worldwide. Owing to the heterogeneity as well as various molecular subtypes, the molecular pathways underlying carcinogenesis in various forms of BC are distinct. Therefore, the advancement of alternative therapy is required to combat the ailment. Recent analyses propose that long non-coding RNAs (lncRNAs) perform an essential function in controlling immune response, and therefore, may provide essential information about the disorder. However, their function in patients with triple-negative BC (TNBC) has not been explored in detail. Here, we analyzed the changes in the genomic expression of messenger RNA (mRNA) and lncRNA in standard control in response to cancer metastasis using publicly available single-cell RNA-Seq data. We identified a total of 197 potentially novel lncRNAs in TNBC patients of which 86 were differentially upregulated and 111 were differentially downregulated. In addition, among the 909 candidate lncRNA transcripts, 19 were significantly differentially expressed (DE) of which three were upregulated and 16 were downregulated. On the other hand, 1901 mRNA transcripts were significantly DE of which 1110 were upregulated and 791 were downregulated by TNBCs subtypes. The Gene Ontology (GO) analyses showed that some of the host genes were enriched in various biological, molecular, and cellular functions. The Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis showed that some of the genes were involved in only one pathway of prostate cancer. The lncRNA-miRNA-gene network analysis showed that the lncRNAs TCONS_00076394 and TCONS_00051377 interacted with breast cancer-related micro RNAs (miRNAs) and the host genes of these lncRNAs were also functionally related to breast cancer. Thus, this study provides novel lncRNAs as potential biomarkers for the therapeutic intervention of this cancer subtype.
Keywords: lncRNA biomarker; long non-coding RNA; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

