D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers
- PMID: 34830296
- PMCID: PMC8625115
- DOI: 10.3390/ijms222212410
D-Xylose Sensing in Saccharomyces cerevisiae: Insights from D-Glucose Signaling and Native D-Xylose Utilizers
Abstract
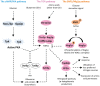
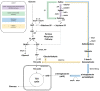
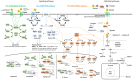
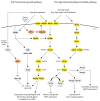
Extension of the substrate range is among one of the metabolic engineering goals for microorganisms used in biotechnological processes because it enables the use of a wide range of raw materials as substrates. One of the most prominent examples is the engineering of baker's yeast Saccharomyces cerevisiae for the utilization of d-xylose, a five-carbon sugar found in high abundance in lignocellulosic biomass and a key substrate to achieve good process economy in chemical production from renewable and non-edible plant feedstocks. Despite many excellent engineering strategies that have allowed recombinant S. cerevisiae to ferment d-xylose to ethanol at high yields, the consumption rate of d-xylose is still significantly lower than that of its preferred sugar d-glucose. In mixed d-glucose/d-xylose cultivations, d-xylose is only utilized after d-glucose depletion, which leads to prolonged process times and added costs. Due to this limitation, the response on d-xylose in the native sugar signaling pathways has emerged as a promising next-level engineering target. Here we review the current status of the knowledge of the response of S. cerevisiae signaling pathways to d-xylose. To do this, we first summarize the response of the native sensing and signaling pathways in S. cerevisiae to d-glucose (the preferred sugar of the yeast). Using the d-glucose case as a point of reference, we then proceed to discuss the known signaling response to d-xylose in S. cerevisiae and current attempts of improving the response by signaling engineering using native targets and synthetic (non-native) regulatory circuits.
Keywords: Saccharomyces cerevisiae; d-xylose; non-native substrate; signaling network engineering; sugar sensing; sugar signaling; synthetic signaling circuits.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

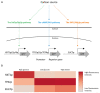
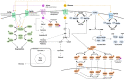
References
-
- Abril M.A., Michan C., Timmis K.N., Ramos J.L. Regulator and Enzyme Specificities of the Tol Plasmid-Encoded Upper Pathway for Degradation of Aromatic-Hydrocarbons and Expansion of the Substrate Range of the Pathway. J. Bacteriol. 1989;171:6782–6790. doi: 10.1128/jb.171.12.6782-6790.1989. - DOI - PMC - PubMed
-
- Klemba M., Jakobs B., Wittich R.M., Pieper D. Chromosomal integration of tcb chlorocatechol degradation pathway genes as a means of expanding the growth substrate range of bacteria to include haloaromatics. Appl. Environ. Microbiol. 2000;66:3255–3261. doi: 10.1128/AEM.66.8.3255-3261.2000. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

