The Large GTPase, GBP-2, Regulates Rho Family GTPases to Inhibit Migration and Invadosome Formation in Breast Cancer Cells
- PMID: 34830789
- PMCID: PMC8616281
- DOI: 10.3390/cancers13225632
The Large GTPase, GBP-2, Regulates Rho Family GTPases to Inhibit Migration and Invadosome Formation in Breast Cancer Cells
Abstract
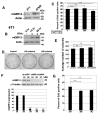
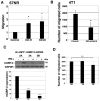
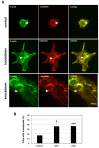
Breast cancer is the most common cancer in women. Despite advances in early detection and treatment, it is predicted that over 43,000 women will die of breast cancer in 2021. To lower this number, more information about the molecular players in breast cancer are needed. Guanylate-Binding Protein-2 has been correlated with better prognosis in breast cancer. In this study, we asked if the expression of GBP-2 in breast cancer merely provided a biomarker for improved prognosis or whether it actually contributed to improving outcome. To answer this, the 4T1 model of murine breast cancer was used. 4T1 cells themselves are highly aggressive and highly metastatic, while 67NR cells, isolated from the same tumor, do not leave the primary site. The expression of GBP-2 was examined in the two cell lines and found to be inversely correlated with aggressiveness/metastasis. Proliferation, migration, and invadosome formation were analyzed after altering the expression levels of GBP-2. Our experiments show that GBP-2 does not alter the proliferation of these cells but inhibits migration and invadosome formation downstream of regulation of Rho GTPases. Together these data demonstrate that GBP-2 is responsible for cell autonomous activities that make breast cancer cells less aggressive.
Keywords: GTPase; Guanylate-Binding Protein; Rho; breast cancer; cytoskeleton; invadosome; migration.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Breastcancer.org. U.S. Breast Cancer Statistics. [(accessed on 3 January 2019)]. Available online: www.breastcancer.org/symptoms/understand_bc/statistics2018.
-
- Lipnik K., Naschberger E., Gonin-Laurent N., Kodajova P., Petznek H., Rungaldier S., Astigiano S., Ferrini S., Sturzl M., Hohenadl C. Interferon g-induced human guanylate-binding protein 1 inhibits mammary tumor growth in mice. Mol. Med. 2010;16:177–187. doi: 10.2119/molmed.2009.00172. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

