Single-Cell Sequencing: Biological Insight and Potential Clinical Implications in Pediatric Leukemia
- PMID: 34830811
- PMCID: PMC8616124
- DOI: 10.3390/cancers13225658
Single-Cell Sequencing: Biological Insight and Potential Clinical Implications in Pediatric Leukemia
Abstract
Single-cell sequencing (SCS) provides high-resolution insight into the genomic, epigenomic, and transcriptomic landscape of oncohematological malignancies including pediatric leukemia, the most common type of childhood cancer. Besides broadening our biological understanding of cellular heterogeneity, sub-clonal architecture, and regulatory network of tumor cell populations, SCS can offer clinically relevant, detailed characterization of distinct compartments affected by leukemia and identify therapeutically exploitable vulnerabilities. In this review, we provide an overview of SCS studies focused on the high-resolution genomic and transcriptomic scrutiny of pediatric leukemia. Our aim is to investigate and summarize how different layers of single-cell omics approaches can expectedly support clinical decision making in the future. Although the clinical management of pediatric leukemia underwent a spectacular improvement during the past decades, resistant disease is a major cause of therapy failure. Currently, only a small proportion of childhood leukemia patients benefit from genomics-driven therapy, as 15-20% of them meet the indication criteria of on-label targeted agents, and their overall response rate falls in a relatively wide range (40-85%). The in-depth scrutiny of various cell populations influencing the development, progression, and treatment resistance of different disease subtypes can potentially uncover a wider range of driver mechanisms for innovative therapeutic interventions.
Keywords: cellular heterogeneity; evolutionary trajectory; pediatric leukemia; single-cell sequencing; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
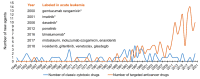

References
-
- Alpar D., Barber L.J., Gerlinger M. Genetic Intratumor Heterogeneity. Epigenetic Cancer Ther. 2015:571–593. doi: 10.1016/B978-0-12-800206-3.00024-0. - DOI
-
- Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A., et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. - DOI - PMC - PubMed
Publication types
Grants and funding
- FK20_134253, K21_137948 and ED18-1-2019-0019/Hungarian National Research, Development and Innovation Office
- 739593/EU's Horizon 2020 research and innovation program
- BO/00320/18/5/János Bolyai Research Scholarship
- ÚNKP-20-5-SE-22, ÚNKP-20-3-II-SE-20 and KDP-2020-1008491/New National Excellence Program and Co-operative Doctoral Program of the Ministry for In-novation and Technology
- 03/MGYH-MGYGYT/2021/Hungarian Pediatric Oncology Network
LinkOut - more resources
Full Text Sources
Miscellaneous

