Biological Significance and Targeting of the FGFR Axis in Cancer
- PMID: 34830836
- PMCID: PMC8616401
- DOI: 10.3390/cancers13225681
Biological Significance and Targeting of the FGFR Axis in Cancer
Abstract
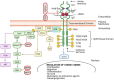
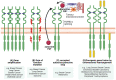
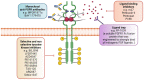
The pleiotropic effects of fibroblast growth factors (FGFs), the widespread expression of all seven signalling FGF receptors (FGFRs) throughout the body, and the dramatic phenotypes shown by many FGF/R knockout mice, highlight the diversity, complexity and functional importance of FGFR signalling. The FGF/R axis is critical during normal tissue development, homeostasis and repair. Therefore, it is not surprising that substantial evidence also pinpoints the involvement of aberrant FGFR signalling in disease, including tumourigenesis. FGFR aberrations in cancer include mutations, gene fusions, and amplifications as well as corrupted autocrine/paracrine loops. Indeed, many clinical trials on cancer are focusing on targeting the FGF/FGFR axis, using selective FGFR inhibitors, nonselective FGFR tyrosine kinase inhibitors, ligand traps, and monoclonal antibodies and some have already been approved for the treatment of cancer patients. The heterogeneous tumour microenvironment and complexity of FGFR signalling may be some of the factors responsible for the resistance or poor response to therapy with FGFR axis-directed therapeutic agents. In the present review we will focus on the structure and function of FGF(R)s, their common irregularities in cancer and the therapeutic value of targeting their function in cancer.
Keywords: FGFR inhibitors; FGFR mutations; FGFR signalling; cancer; fibroblast growth factor; targeting FGFR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

