Inhibitors of the Plasmodium falciparum Hsp90 towards Selective Antimalarial Drug Design: The Past, Present and Future
- PMID: 34831072
- PMCID: PMC8616389
- DOI: 10.3390/cells10112849
Inhibitors of the Plasmodium falciparum Hsp90 towards Selective Antimalarial Drug Design: The Past, Present and Future
Abstract
Malaria is still one of the major killer parasitic diseases in tropical settings, posing a public health threat. The development of antimalarial drug resistance is reversing the gains made in attempts to control the disease. The parasite leads a complex life cycle that has adapted to outwit almost all known antimalarial drugs to date, including the first line of treatment, artesunate. There is a high unmet need to develop new strategies and identify novel therapeutics to reverse antimalarial drug resistance development. Among the strategies, here we focus and discuss the merits of the development of antimalarials targeting the Heat shock protein 90 (Hsp90) due to the central role it plays in protein quality control.
Keywords: Hsp90 inhibitors; Plasmodium falciparum; heat shock protein 90; malaria; molecular chaperone; parasite Hsp90.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



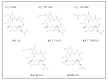
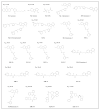
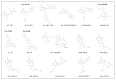

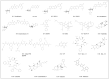
References
-
- World Health Organisation World Malaria Report, 2020. [(accessed on 23 September 2021)]. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria....
-
- Warrell D.A., Gilles H.M. Essential Malariology. 4th ed. CRC Press; Boca Raton, FL, USA: 2002.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

