Changes in Cell Morphology and Actin Organization in Embryonic Stem Cells Cultured under Different Conditions
- PMID: 34831083
- PMCID: PMC8616278
- DOI: 10.3390/cells10112859
Changes in Cell Morphology and Actin Organization in Embryonic Stem Cells Cultured under Different Conditions
Abstract
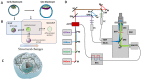
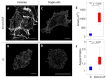
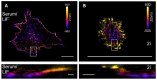
The cellular cytoskeleton provides the cell with a mechanical rigidity that allows mechanical interaction between cells and the extracellular environment. The actin structure plays a key role in mechanical events such as motility or the establishment of cell polarity. From the earliest stages of development, as represented by the ex vivo expansion of naïve embryonic stem cells (ESCs), the critical mechanical role of the actin structure is becoming recognized as a vital cue for correct segregation and lineage control of cells and as a regulatory structure that controls several transcription factors. Naïve ESCs have a characteristic morphology, and the ultrastructure that underlies this condition remains to be further investigated. Here, we investigate the 3D actin cytoskeleton of naïve mouse ESCs using super-resolution optical reconstruction microscopy (STORM). We investigate the morphological, cytoskeletal, and mechanical changes in cells cultured in 2i or Serum/LIF media reflecting, respectively, a homogeneous preimplantation cell state and a state that is closer to embarking on differentiation. STORM imaging showed that the peripheral actin structure undergoes a dramatic change between the two culturing conditions. We also detected micro-rheological differences in the cell periphery between the cells cultured in these two media correlating well with the observed nano-architecture of the ESCs in the two different culture conditions. These results pave the way for linking physical properties and cytoskeletal architecture to cell morphology during early development.
Keywords: actin cytoskeleton; cell culturing; embryonic stem cells; micro-rheology; optical tweezers; primed embryonic stem cells; super-resolution microscopy (STORM).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bongiorno T., Gura J., Talwar P., Chambers D., Young K.M., Arafat D., Wang G., Jackson-Holmes E.L., Qiu P., McDevitt T.C., et al. Biophysical subsets of embryonic stem cells display distinct phenotypic and morphological signatures. PLoS ONE. 2018;13:e0192631. doi: 10.1371/journal.pone.0192631. - DOI - PMC - PubMed
-
- Urbanska M., Winzi M., Neumann K., Abuhattum S., Rosendahl P., Muller P., Taubenberger A., Anastassiadis K., Guck J. Single-cell mechanical phenotype is an intrinsic marker of reprogramming and differentiation along the mouse neural lineage. Development. 2017;144:4313–4321. doi: 10.1242/dev.155218. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

