Molecular Mechanisms of mtDNA-Mediated Inflammation
- PMID: 34831121
- PMCID: PMC8616383
- DOI: 10.3390/cells10112898
Molecular Mechanisms of mtDNA-Mediated Inflammation
Abstract
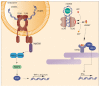
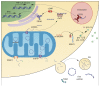
Besides their role in cell metabolism, mitochondria display many other functions. Mitochondrial DNA (mtDNA), the own genome of the organelle, plays an important role in modulating the inflammatory immune response. When released from the mitochondrion to the cytosol, mtDNA is recognized by cGAS, a cGAMP which activates a pathway leading to enhanced expression of type I interferons, and by NLRP3 inflammasome, which promotes the activation of pro-inflammatory cytokines Interleukin-1beta and Interleukin-18. Furthermore, mtDNA can be bound by Toll-like receptor 9 in the endosome and activate a pathway that ultimately leads to the expression of pro-inflammatory cytokines. mtDNA is released in the extracellular space in different forms (free DNA, protein-bound DNA fragments) either as free circulating molecules or encapsulated in extracellular vesicles. In this review, we discussed the latest findings concerning the molecular mechanisms that regulate the release of mtDNA from mitochondria, and the mechanisms that connect mtDNA misplacement to the activation of inflammation in different pathophysiological conditions.
Keywords: STING; TLR9; extracellular cf-mtDNA; inflammasome; mitochondria; mtDNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

