DNA Methylation and Immune Memory Response
- PMID: 34831166
- PMCID: PMC8616503
- DOI: 10.3390/cells10112943
DNA Methylation and Immune Memory Response
Abstract
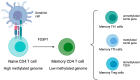
The generation of memory is a cardinal feature of the adaptive immune response, involving different factors in a complex process of cellular differentiation. This process is essential for protecting the second encounter with pathogens and is the mechanism by which vaccines work. Epigenetic changes play important roles in the regulation of cell differentiation events. There are three types of epigenetic regulation: DNA methylation, histone modification, and microRNA expression. One of these epigenetic changes, DNA methylation, occurs in cytosine residues, mainly in CpG dinucleotides. This brief review aimed to analyse the literature to verify the involvement of DNA methylation during memory T and B cell development. Several studies have highlighted the importance of the DNA methyltransferases, enzymes that catalyse the methylation of DNA, during memory differentiation, maintenance, and function. The methylation profile within different subsets of naïve activated and memory cells could be an interesting tool to help monitor immune memory response.
Keywords: B cells; DNA methylation; T cells; epigenetic; memory response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

