RALBP1 in Oxidative Stress and Mitochondrial Dysfunction in Alzheimer's Disease
- PMID: 34831336
- PMCID: PMC8620796
- DOI: 10.3390/cells10113113
RALBP1 in Oxidative Stress and Mitochondrial Dysfunction in Alzheimer's Disease
Abstract
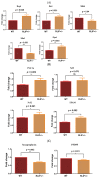
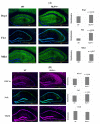
The purpose of our study is to understand the role of the RALBP1 gene in oxidative stress (OS), mitochondrial dysfunction and cognition in Alzheimer's disease (AD) pathogenesis. The RALPB1 gene encodes the 76 kDa protein RLIP76 (Rlip). Rlip functions as a stress-responsive/protective transporter of glutathione conjugates (GS-E) and xenobiotic toxins. We hypothesized that Rlip may play an important role in maintaining cognitive function. The aim of this study is to determine whether Rlip deficiency in mice is associated with AD-like cognitive and mitochondrial dysfunction. Brain tissue obtained from cohorts of wildtype (WT) and Rlip+/- mice were analyzed for OS markers, expression of genes that regulate mitochondrial fission/fusion, and synaptic integrity. We also examined mitochondrial ultrastructure in brains obtained from these mice and further analyzed the impact of Rlip deficiency on gene networks of AD, aging, stress response, mitochondrial function, and CREB signaling. Our studies revealed a significant increase in the levels of OS markers and alterations in the expression of genes and proteins involved in mitochondrial biogenesis, dynamics and synapses in brain tissues from these mice. Furthermore, we compared the cognitive function of WT and Rlip+/- mice. Behavioral, basic motor and sensory function tests in Rlip+/- mice revealed cognitive decline, similar to AD. Gene network analysis indicated dysregulation of stress-activated gene expression, mitochondrial function and CREB signaling genes in the Rlip+/- mouse brain. Our results suggest that Rlip deficiency-associated increases in OS and mitochondrial dysfunction could contribute to the development or progression of OS-related AD processes.
Keywords: Alzheimer’s disease; mitochondria; mitochondrial biogenesis; mitophagy; synaptic proteins.
Conflict of interest statement
The authors, except S.A., do not have any competing financial interests that could influence, or give the perception of such influence on, the behavior or content in a way that could undermine the objectivity, integrity or perceived value of this publication. S.A. is a founder of AVESTA76 Therapeutics, which will develop Rlip-targeting small molecules for therapy.
Figures









References
-
- Knopman D.S., DeKosky S.T., Cummings J.L., Chui H., Corey-Bloom J., Relkin N., Small G.W., Miller B., Stevens J.C. Practice parameter: Diagnosis of dementia (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2001;56:1143–1153. doi: 10.1212/WNL.56.9.1143. - DOI - PubMed
-
- Mormino E.C., Betensky R.A., Hedden T., Schultz A.P., Amariglio R.E., Rentz D.M., Johnson K.A., Sperling R.A. Synergistic effect of β-amyloid and neurodegeneration on cognitive decline in clinically normal individuals. JAMA Neurol. 2014;71:1379–1385. doi: 10.1001/jamaneurol.2014.2031. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

