Babesia microti Immunoreactive Rhoptry-Associated Protein-1 Paralogs Are Ancestral Members of the Piroplasmid-Confined RAP-1 Family
- PMID: 34832541
- PMCID: PMC8624774
- DOI: 10.3390/pathogens10111384
Babesia microti Immunoreactive Rhoptry-Associated Protein-1 Paralogs Are Ancestral Members of the Piroplasmid-Confined RAP-1 Family
Abstract
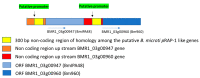
Babesia, Cytauxzoon and Theileria are tick-borne apicomplexan parasites of the order Piroplasmida, responsible for diseases in humans and animals. Members of the piroplasmid rhoptry-associated protein-1 (pRAP-1) family have a signature cysteine-rich domain and are important for parasite development. We propose that the closely linked B. microti genes annotated as BMR1_03g00947 and BMR1_03g00960 encode two paralogue pRAP-1-like proteins named BmIPA48 and Bm960. The two genes are tandemly arranged head to tail, highly expressed in blood stage parasites, syntenic to rap-1 genes of other piroplasmids, and share large portions of an almost identical ~225 bp sequence located in their 5' putative regulatory regions. BmIPA48 and Bm960 proteins contain a N-terminal signal peptide, share very low sequence identity (<13%) with pRAP-1 from other species, and harbor one or more transmembrane domains. Diversification of the piroplasmid-confined prap-1 family is characterized by amplification of genes, protein domains, and a high sequence polymorphism. This suggests a functional involvement of pRAP-1 at the parasite-host interface, possibly in parasite adhesion, attachment, and/or evasion of the host immune defenses. Both BmIPA48 and Bm960 are recognized by antibodies in sera from humans infected with B. microti and might be promising candidates for developing novel serodiagnosis and vaccines.
Keywords: BMR1_03g00960; Babesia microti; BmIPA48; human babesiosis; piroplasmid rhoptry-associated protein-1 (pRAP-1).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

