HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening
- PMID: 34832897
- PMCID: PMC8621417
- DOI: 10.3390/ph14111115
HIV-1 Protease and Reverse Transcriptase Inhibitory Activities of Curcuma aeruginosa Roxb. Rhizome Extracts and the Phytochemical Profile Analysis: In Vitro and In Silico Screening
Abstract
Human immunodeficiency virus type-1 (HIV-1) infection causes acquired immunodeficiency syndrome (AIDS). Currently, several anti-retroviral drugs are available, but adverse effects of these drugs have been reported. Herein, we focused on the anti-HIV-1 activity of Curcuma aeruginosa Roxb. (CA) extracted by hexane (CA-H), ethyl acetate (CA-EA), and methanol (CA-M). The in vitro HIV-1 protease (PR) and HIV-1 reverse transcriptase (RT) inhibitory activities of CA extracts were screened. CA-M potentially inhibited HIV-1 PR (82.44%) comparable to Pepstatin A (81.48%), followed by CA-EA (67.05%) and CA-H (47.6%), respectively. All extracts exhibited moderate inhibition of HIV-1 RT (64.97 to 76.93%). Besides, phytochemical constituents of CA extracts were identified by GC-MS and UPLC-HRMS. Fatty acids, amino acids, and terpenoids were the major compounds found in the extracts. Furthermore, drug-likeness parameters and the ability of CA-identified compounds on blocking of the HIV-1 PR and RT active sites were in silico investigated. Dihydroergocornine, 3β,6α,7α-trihydroxy-5β-cholan-24-oic acid, and 6β,11β,16α,17α,21-Pentahydroxypregna-1,4-diene-3,20-dione-16,17-acetonide showed strong binding affinities at the active residues of both HIV-1 PR and RT. Moreover, antioxidant activity of CA extracts was determined. CA-EA exhibited the highest antioxidant activity, which positively related to the amount of total phenolic content. This study provided beneficial data for anti-HIV-1 drug discovery from CA extracts.
Keywords: ADMET analysis; AIDS; Curcuma aeruginosa Roxb.; HIV-1 protease inhibitor; HIV-1 reverse transcriptase inhibitor; drug discovery; immunodeficiency virus; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

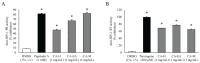





References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

