Understanding the Mechanism of Action of NAI-112, a Lanthipeptide with Potent Antinociceptive Activity
- PMID: 34833857
- PMCID: PMC8624038
- DOI: 10.3390/molecules26226764
Understanding the Mechanism of Action of NAI-112, a Lanthipeptide with Potent Antinociceptive Activity
Abstract
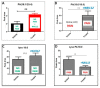
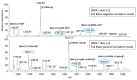
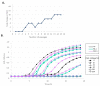
NAI-112, a glycosylated, labionine-containing lanthipeptide with weak antibacterial activity, has demonstrated analgesic activity in relevant mouse models of nociceptive and neuropathic pain. However, the mechanism(s) through which NAI-112 exerts its analgesic and antibacterial activities is not known. In this study, we analyzed changes in the spinal cord lipidome resulting from treatment with NAI-112 of naive and in-pain mice. Notably, NAI-112 led to an increase in phosphatidic acid levels in both no-pain and pain models and to a decrease in lysophosphatidic acid levels in the pain model only. We also showed that NAI-112 can form complexes with dipalmitoyl-phosphatidic acid and that Staphylococcus aureus can become resistant to NAI-112 through serial passages at sub-inhibitory concentrations of the compound. The resulting resistant mutants were phenotypically and genotypically related to vancomycin-insensitive S. aureus strains, suggesting that NAI-112 binds to the peptidoglycan intermediate lipid II. Altogether, our results suggest that NAI-112 binds to phosphate-containing lipids and blocks pain sensation by decreasing levels of lysophosphatidic acid in the TRPV1 pathway.
Keywords: TPRV1; VISA strains; lanthipeptide; lipid II; lysophosphatidic acid; untargeted lipidomics.
Conflict of interest statement
The authors declare the following competing financial interest: M.I., A.R., and S.D. are inventors of the patent on NAI-112 owned by NAICONS and IIT. A.T., M.I. and S.D. are employees and shareholders of NAICONS.
Figures



References
-
- Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J., et al. Ribosomally Synthesized and Post-Translationally Modified Peptide Natural Products: Overview and Recommendations for a Universal Nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

