Virtual Screening for Potential Phytobioactives as Therapeutic Leads to Inhibit NQO1 for Selective Anticancer Therapy
- PMID: 34833955
- PMCID: PMC8622762
- DOI: 10.3390/molecules26226863
Virtual Screening for Potential Phytobioactives as Therapeutic Leads to Inhibit NQO1 for Selective Anticancer Therapy
Abstract
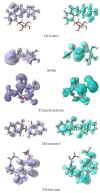
NAD(P)H:quinone acceptor oxidoreductase-1 (NQO1) is a ubiquitous flavin adenine dinucleotide-dependent flavoprotein that promotes obligatory two-electron reductions of quinones, quinonimines, nitroaromatics, and azo dyes. NQO1 is a multifunctional antioxidant enzyme whose expression and deletion are linked to reduced and increased oxidative stress susceptibilities. NQO1 acts as both a tumor suppressor and tumor promoter; thus, the inhibition of NQO1 results in less tumor burden. In addition, the high expression of NQO1 is associated with a shorter survival time of cancer patients. Inhibiting NQO1 also enables certain anticancer agents to evade the detoxification process. In this study, a series of phytobioactives were screened based on their chemical classes such as coumarins, flavonoids, and triterpenoids for their action on NQO1. The in silico evaluations were conducted using PyRx virtual screening tools, where the flavone compound, Orientin showed a better binding affinity score of -8.18 when compared with standard inhibitor Dicumarol with favorable ADME properties. An MD simulation study found that the Orientin binding to NQO1 away from the substrate-binding site induces a potential conformational change in the substrate-binding site, thereby inhibiting substrate accessibility towards the FAD-binding domain. Furthermore, with this computational approach we are offering a scope for validation of the new therapeutic components for their in vitro and in vivo efficacy against NQO1.
Keywords: NQO1; Nrf2; binding affinity; conceptual DFT; detoxification; molecular docking; phytobiactives; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

