A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19
- PMID: 34834174
- PMCID: PMC8624722
- DOI: 10.3390/pharmaceutics13111759
A Comprehensive Review about the Molecular Structure of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): Insights into Natural Products against COVID-19
Abstract
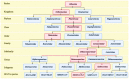
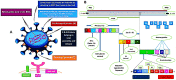
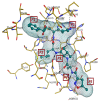
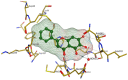
In 2019, the world suffered from the emergence of COVID-19 infection, one of the most difficult pandemics in recent history. Millions of confirmed deaths from this pandemic have been reported worldwide. This disaster was caused by SARS-CoV-2, which is the last discovered member of the family of Coronaviridae. Various studies have shown that natural compounds have effective antiviral properties against coronaviruses by inhibiting multiple viral targets, including spike proteins and viral enzymes. This review presents the classification and a detailed explanation of the SARS-CoV-2 molecular characteristics and structure-function relationships. We present all currently available crystal structures of different SARS-CoV-2 proteins and emphasized on the crystal structure of different virus proteins and the binding modes of their ligands. This review also discusses the various therapeutic approaches for COVID-19 treatment and available vaccinations. In addition, we highlight and compare the existing data about natural compounds extracted from algae, fungi, plants, and scorpion venom that were used as antiviral agents against SARS-CoV-2 infection. Moreover, we discuss the repurposing of select approved therapeutic agents that have been used in the treatment of other viruses.
Keywords: COVID-19; SARS-CoV-2; antioxidants; antivirals; coronavirus; molecular structure; natural products; therapeutic approach; vaccines; virus detection; virus lifecycle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

