A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant
- PMID: 34834827
- PMCID: PMC8624978
- DOI: 10.3390/plants10112464
A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant
Abstract
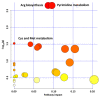
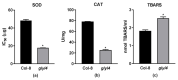
Methylglyoxal (MG) is a cytotoxic compound often produced as a side product of metabolic processes such as glycolysis, lipid peroxidation, and photosynthesis. MG is mainly scavenged by the glyoxalase system, a two-step pathway, in which the coordinate activity of GLYI and GLYII transforms it into D-lactate, releasing GSH. In Arabidopsis thaliana, a member of the GLYI family named GLYI4 has been recently characterized. In glyI4 mutant plants, a general stress phenotype characterized by compromised MG scavenging, accumulation of reactive oxygen species (ROS), stomatal closure, and reduced fitness was observed. In order to shed some light on the impact of gly4 loss-of-function on plant metabolism, we applied a high resolution mass spectrometry-based metabolomic approach to Arabidopsis Col-8 wild type and glyI4 mutant plants. A compound library containing a total of 70 metabolites, differentially synthesized in glyI4 compared to Col-8, was obtained. Pathway analysis of the identified compounds showed that the upregulated pathways are mainly involved in redox reactions and cellular energy maintenance, and those downregulated in plant defense and growth. These results improved our understanding of the impacts of glyI4 loss-of-function on the general reprogramming of the plant's metabolic landscape as a strategy for surviving under adverse physiological conditions.
Keywords: glyoxalase I; metabolite profiling; methylglyoxal; oxidative stress; plant defense; plant growth.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

