Interstitial Telomeric-like Repeats (ITR) in Seed Plants as Assessed by Molecular Cytogenetic Techniques: A Review
- PMID: 34834904
- PMCID: PMC8621592
- DOI: 10.3390/plants10112541
Interstitial Telomeric-like Repeats (ITR) in Seed Plants as Assessed by Molecular Cytogenetic Techniques: A Review
Abstract
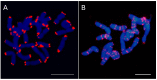
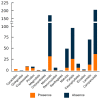
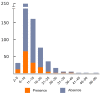
The discovery of telomeric repeats in interstitial regions of plant chromosomes (ITRs) through molecular cytogenetic techniques was achieved several decades ago. However, the information is scattered and has not been critically evaluated from an evolutionary perspective. Based on the analysis of currently available data, it is shown that ITRs are widespread in major evolutionary lineages sampled. However, their presence has been detected in only 45.6% of the analysed families, 26.7% of the sampled genera, and in 23.8% of the studied species. The number of ITR sites greatly varies among congeneric species and higher taxonomic units, and range from one to 72 signals. ITR signals mostly occurs as homozygous loci in most species, however, odd numbers of ITR sites reflecting a hemizygous state have been reported in both gymnosperm and angiosperm groups. Overall, the presence of ITRs appears to be poor predictors of phylogenetic and taxonomic relatedness at most hierarchical levels. The presence of ITRs and the number of sites are not significantly associated to the number of chromosomes. The longitudinal distribution of ITR sites along the chromosome arms indicates that more than half of the ITR presences are between proximal and terminal locations (49.5%), followed by proximal (29.0%) and centromeric (21.5%) arm regions. Intraspecific variation concerning ITR site number, chromosomal locations, and the differential presence on homologous chromosome pairs has been reported in unrelated groups, even at the population level. This hypervariability and dynamism may have likely been overlooked in many lineages due to the very low sample sizes often used in cytogenetic studies.
Keywords: chromosomal landmarks; in situ hybridisation; interstitial telomeric repeats; karyological evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Inter- and intraspecific hypervariability in interstitial telomeric-like repeats (TTTAGGG)n in Anacyclus (Asteraceae).Ann Bot. 2018 Aug 27;122(3):387-395. doi: 10.1093/aob/mcy079. Ann Bot. 2018. PMID: 29800070 Free PMC article.
-
Interstitial Arabidopsis-Type Telomeric Repeats in Asteraceae.Plants (Basel). 2021 Dec 17;10(12):2794. doi: 10.3390/plants10122794. Plants (Basel). 2021. PMID: 34961265 Free PMC article.
-
Interstitial telomeric repeats are enriched in the centromeres of chromosomes in Solanum species.Chromosome Res. 2013 Mar;21(1):5-13. doi: 10.1007/s10577-012-9332-x. Epub 2012 Dec 19. Chromosome Res. 2013. PMID: 23250588
-
Interstitial telomeric sites and Robertsonian translocations in species of Ipheion and Nothoscordum (Amaryllidaceae).Genetica. 2016 Apr;144(2):157-66. doi: 10.1007/s10709-016-9886-1. Epub 2016 Feb 11. Genetica. 2016. PMID: 26869260 Review.
-
Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution.Mutat Res Rev Mutat Res. 2017 Jul;773:51-65. doi: 10.1016/j.mrrev.2017.04.002. Epub 2017 Apr 22. Mutat Res Rev Mutat Res. 2017. PMID: 28927537 Review.
Cited by
-
Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View.Plants (Basel). 2022 Mar 16;11(6):784. doi: 10.3390/plants11060784. Plants (Basel). 2022. PMID: 35336666 Free PMC article. Review.
-
Chromosomal dynamics in Senna: comparative PLOP-FISH analysis of tandem repeats and flow cytometric nuclear genome size estimations.Front Plant Sci. 2023 Dec 14;14:1288220. doi: 10.3389/fpls.2023.1288220. eCollection 2023. Front Plant Sci. 2023. PMID: 38173930 Free PMC article.
-
RepeatOBserver: Tandem Repeat Visualisation and Putative Centromere Detection.Mol Ecol Resour. 2025 Oct;25(7):e14084. doi: 10.1111/1755-0998.14084. Epub 2025 Mar 4. Mol Ecol Resour. 2025. PMID: 40035343 Free PMC article.
-
The genome of Citrus australasica reveals disease resistance and other species specific genes.BMC Plant Biol. 2024 Apr 10;24(1):260. doi: 10.1186/s12870-024-04988-8. BMC Plant Biol. 2024. PMID: 38594608 Free PMC article.
-
FISH Mapping of Telomeric and Non-Telomeric (AG3T3)3 Reveal the Chromosome Numbers and Chromosome Rearrangements of 41 Woody Plants.Genes (Basel). 2022 Jul 14;13(7):1239. doi: 10.3390/genes13071239. Genes (Basel). 2022. PMID: 35886022 Free PMC article.
References
-
- Nelson A.D., Beilstein M.A., Shippen D.E. Plant telomeres and telomerase. In: Howell S.H., editor. Molecular Biology. Springer; New York, NY, USA: 2014. pp. 25–49.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

