Mutational Hotspot in the SARS-CoV-2 Spike Protein N-Terminal Domain Conferring Immune Escape Potential
- PMID: 34834921
- PMCID: PMC8618472
- DOI: 10.3390/v13112114
Mutational Hotspot in the SARS-CoV-2 Spike Protein N-Terminal Domain Conferring Immune Escape Potential
Abstract
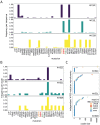
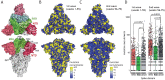
Global efforts are being made to monitor the evolution of SARS-CoV-2, aiming for early identification of genotypes providing increased infectivity or virulence. However, viral lineage-focused tracking might fail in early detection of advantageous mutations emerging independently across phylogenies. Here, the emergence patterns of Spike mutations were investigated in sequences deposited in local and global databases to identify mutational hotspots across phylogenies and we evaluated their impact on SARS-CoV-2 evolution. We found a striking increase in the frequency of recruitment of diverse substitutions at a critical residue (W152), positioned in the N-terminal domain (NTD) of the Spike protein, observed repeatedly across independent phylogenetic and geographical contexts. These mutations might have an impact on the evasion of neutralizing antibodies. Finally, we found that NTD is a region exhibiting particularly high frequency of mutation recruitments, suggesting an evolutionary path in which the virus maintains optimal efficiency of ACE2 binding combined with the flexibility facilitating the immune escape. We conclude that adaptive mutations, frequently present outside of the receptor-binding domain, can emerge in virtually any SARS-CoV-2 lineage and at any geographical location. Therefore, surveillance should not be restricted to monitoring defined lineages alone.
Keywords: SARS-CoV-2 genome; W152; coronavirus; immune escape; neutralizing antibody; spike NTD; viral evolution.
Conflict of interest statement
All authors are employees of SOPHiA GENETICS.
Figures


References
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain That Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

