PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods
- PMID: 34834927
- PMCID: PMC8623551
- DOI: 10.3390/v13112120
PHIDA: A High Throughput Turbidimetric Data Analytic Tool to Compare Host Range Profiles of Bacteriophages Isolated Using Different Enrichment Methods
Abstract
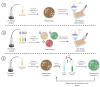
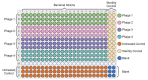
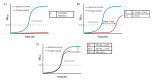
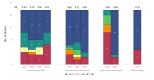
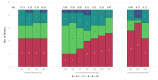
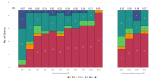
Bacteriophages are viruses that infect bacteria and are present in niches where bacteria thrive. In recent years, the suggested application areas of lytic bacteriophage have been expanded to include therapy, biocontrol, detection, sanitation, and remediation. However, phage application is constrained by the phage's host range-the range of bacterial hosts sensitive to the phage and the degree of infection. Even though phage isolation and enrichment techniques are straightforward protocols, the correlation between the enrichment technique and host range profile has not been evaluated. Agar-based methods such as spotting assay and efficiency of plaquing (EOP) are the most used methods to determine the phage host range. These methods, aside from being labor intensive, can lead to subjective and incomplete results as they rely on qualitative observations of the lysis/plaques, do not reflect the lytic activity in liquid culture, and can overestimate the host range. In this study, phages against three bacterial genera were isolated using three different enrichment methods. Host range profiles of the isolated phages were quantitatively determined using a high throughput turbidimetric protocol and the data were analyzed with an accessible analytic tool "PHIDA". Using this tool, the host ranges of 9 Listeria, 14 Salmonella, and 20 Pseudomonas phages isolated with different enrichment methods were quantitatively compared. A high variability in the host range index (HRi) ranging from 0.86-0.63, 0.07-0.24, and 0.00-0.67 for Listeria, Salmonella, and Pseudomonas phages, respectively, was observed. Overall, no direct correlation was found between the phage host range breadth and the enrichment method in any of the three target bacterial genera. The high throughput method and analytics tool developed in this study can be easily adapted to any phage study and can provide a consensus for phage host range determination.
Keywords: bacteriophage; host range; isolation protocol; lytic phages; phage biocontrol; phage enrichment; phage therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

