Genotypic Variants of Pandemic H1N1 Influenza A Viruses Isolated from Severe Acute Respiratory Infections in Ukraine during the 2015/16 Influenza Season
- PMID: 34834932
- PMCID: PMC8619959
- DOI: 10.3390/v13112125
Genotypic Variants of Pandemic H1N1 Influenza A Viruses Isolated from Severe Acute Respiratory Infections in Ukraine during the 2015/16 Influenza Season
Abstract
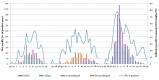
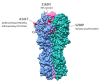
Human type A influenza viruses A(H1N1)pdm09 have caused seasonal epidemics of influenza since the 2009-2010 pandemic. A(H1N1)pdm09 viruses had a leading role in the severe epidemic season of 2015/16 in the Northern Hemisphere and caused a high incidence of acute respiratory infection (ARI) in Ukraine. Serious complications of influenza-associated severe ARI (SARI) were observed in the very young and individuals at increased risk, and 391 fatal cases occurred in the 2015/16 epidemic season. We analyzed the genetic changes in the genomes of A(H1N1)pdm09 influenza viruses isolated from SARI cases in Ukraine during the 2015/16 season. The viral hemagglutinin (HA) fell in H1 group 6B.1 for all but four isolates, with known mutations affecting glycosylation, the Sa antigenic site (S162N in all 6B.1 isolates), or virulence (D222G/N in two isolates). Other mutations occurred in antigenic site Ca (A141P and S236P), and a subgroup of four strains were in group 6B.2, with potential alterations to antigenicity in A(H1N1)pdm09 viruses circulating in 2015/16 in Ukraine. A cluster of Ukrainian isolates exhibited novel D2E and N48S mutations in the RNA binding domain, and E125D in the effector domain, of immune evasion nonstructural protein 1 (NS1). The diverse spectrum of amino-acid substitutions in HA, NS1, and other viral proteins including nucleoprotein (NP) and the polymerase complex suggested the concurrent circulation of multiple lineages of A(H1N1)pdm09 influenza viruses in the human population in Ukraine, a country with low vaccination coverage, complicating public health measures against influenza.
Keywords: A(H1N1)pdm09; H1N1; acute respiratory infection; influenza; mutation; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Belanov S., Bychkov D., Benner C., Ripatti S., Ojala T., Kankainen M., Lee H., Tang J., Kainov D. Genome-Wide Analysis of Evolutionary Markers of Human Influenza A(H1N1)pdm09 and A(H3N2) Viruses May Guide Selection of Vaccine Strain Candidates. Genome Biol. Evol. 2015;7:3472–3483. doi: 10.1093/gbe/evv240. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

