Evaluating RNA Structural Flexibility: Viruses Lead the Way
- PMID: 34834937
- PMCID: PMC8624864
- DOI: 10.3390/v13112130
Evaluating RNA Structural Flexibility: Viruses Lead the Way
Abstract
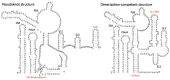
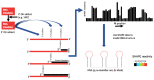
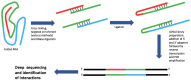
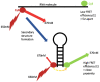
Our understanding of RNA structure has lagged behind that of proteins and most other biological polymers, largely because of its ability to adopt multiple, and often very different, functional conformations within a single molecule. Flexibility and multifunctionality appear to be its hallmarks. Conventional biochemical and biophysical techniques all have limitations in solving RNA structure and to address this in recent years we have seen the emergence of a wide diversity of techniques applied to RNA structural analysis and an accompanying appreciation of its ubiquity and versatility. Viral RNA is a particularly productive area to study in that this economy of function within a single molecule admirably suits the minimalist lifestyle of viruses. Here, we review the major techniques that are being used to elucidate RNA conformational flexibility and exemplify how the structure and function are, as in all biology, tightly linked.
Keywords: NMR; RNA flexibility; RNA structure; RNA viruses; SAXS; SHAPE; proximity ligation; smFRET.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

