Evaluating Large Spontaneous Deletions in a Bovine Cell Line Selected for Bovine Viral Diarrhea Virus Resistance
- PMID: 34834954
- PMCID: PMC8622392
- DOI: 10.3390/v13112147
Evaluating Large Spontaneous Deletions in a Bovine Cell Line Selected for Bovine Viral Diarrhea Virus Resistance
Abstract
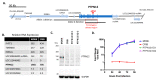
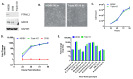
Bovine viral diarrhea virus's (BVDV) entry into bovine cells involves attachment of virions to cellular receptors, internalization, and pH-dependent fusion with endosomal membranes. The primary host receptor for BVDV is CD46; however, the complete set of host factors required for virus entry is unknown. The Madin-Darby bovine kidney (MDBK) cell line is susceptible to BVDV infection, while a derivative cell line (CRIB) is resistant at the level of virus entry. We performed complete genome sequencing of each to identify genomic variation underlying the resistant phenotype with the aim of identifying host factors essential for BVDV entry. Three large compound deletions in the BVDV-resistant CRIB cell line were identified and predicted to disrupt the function or expression of the genes PTPN12, GRID2, and RABGAP1L. However, CRISPR/Cas9 mediated knockout of these genes, individually or in combination, in the parental MDBK cell line did not impact virus entry or replication. Therefore, resistance to BVDV in the CRIB cell line is not due to the apparent spontaneous loss of PTPN12, GRID2, or RABGAP1L gene function. Identifying the functional cause of BVDV resistance in the CRIB cell line may require more detailed comparisons of the genomes and epigenomes.
Keywords: BVDV; CRIB; CRISPR; GRID2; MDBK; PTPN12; RABGAP1L.
Conflict of interest statement
D.A.W. and D.F.C. are full time employees of Recombinetics, Inc. and T. S. S. is an employee of Acceligen, a subsidiary of Recombinetics, Inc. Recombinetics, Inc. is a company that commercializes animal gene editing and associated applied technologies for biomedical research, regenerative medicine and animal agriculture. There are no patents to declare and the interests do not alter the authors’ adherence to all the journal’s policies on sharing data and materials published herein.
Figures



References
-
- Olafson P., Maccallum A.D., Fox F.H. An apparently new transmissible disease of cattle. Cornell Vet. 1946;36:205–213. - PubMed
-
- Walz P.H., Chamorro M.F., Falkenberg M.S., Passler T., van der Meer F., Woolums R.A. Bovine viral diarrhea virus: An updated American College of Veterinary Internal Medicine consensus statement with focus on virus biology, hosts, immunosuppression, and vaccination. J. Vet. Intern. Med. 2020;34:1690–1706. doi: 10.1111/jvim.15816. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

