Interspecies Jumping of Bat Coronaviruses
- PMID: 34834994
- PMCID: PMC8620431
- DOI: 10.3390/v13112188
Interspecies Jumping of Bat Coronaviruses
Abstract
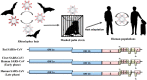
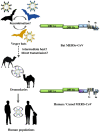
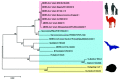
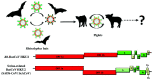
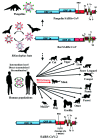
In the last two decades, several coronavirus (CoV) interspecies jumping events have occurred between bats and other animals/humans, leading to major epidemics/pandemics and high fatalities. The SARS epidemic in 2002/2003 had a ~10% fatality. The discovery of SARS-related CoVs in horseshoe bats and civets and genomic studies have confirmed bat-to-civet-to-human transmission. The MERS epidemic that emerged in 2012 had a ~35% mortality, with dromedaries as the reservoir. Although CoVs with the same genome organization (e.g., Tylonycteris BatCoV HKU4 and Pipistrellus BatCoV HKU5) were also detected in bats, there is still a phylogenetic gap between these bat CoVs and MERS-CoV. In 2016, 10 years after the discovery of Rhinolophus BatCoV HKU2 in Chinese horseshoe bats, fatal swine disease outbreaks caused by this virus were reported in southern China. In late 2019, an outbreak of pneumonia emerged in Wuhan, China, and rapidly spread globally, leading to >4,000,000 fatalities so far. Although the genome of SARS-CoV-2 is highly similar to that of SARS-CoV, patient zero and the original source of the pandemic are still unknown. To protect humans from future public health threats, measures should be taken to monitor and reduce the chance of interspecies jumping events, either occurring naturally or through recombineering experiments.
Keywords: COVID-19; MERS; SADS; SARS; bat; coronavirus; epidemic; interspecies jumping; outbreak; pandemic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- ICTV Taxonomy History: Cornidovirineae. [(accessed on 2 January 2019)]. Available online: https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=20186105.
-
- Lau S.K.P., Wong A.C.P., Zhang L., Luk H.K.H., Kwok J.S.L., Ahmed S.S., Cai J.P., Zhao P.S.H., Teng J.L.L., Tsui S.K.W., et al. Novel Bat Alphacoronaviruses in Southern China Support Chinese Horseshoe Bats as an Important Reservoir for Potential Novel Coronaviruses. Viruses. 2019;11:423. doi: 10.3390/v11050423. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- T11/707/15/Theme-based Research Scheme, University Grant Committee
- Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health, HKSAR Government
- CID-HKU1-6/Health and Medical Research Fund of the Food and Health Bureau of HKSAR
- COVID190122/Health and Medical Research Fund of the Food and Health Bureau of HKSAR
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

